Porphyromonas gingivalis-mediated Epithelial Cell Entry of HIV-1
- PMID: 24874702
- PMCID: PMC4126220
- DOI: 10.1177/0022034514537647
Porphyromonas gingivalis-mediated Epithelial Cell Entry of HIV-1
Abstract
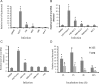
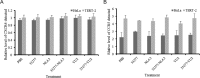
HIV-1 relies on the host's cell machinery to establish a successful infection. Surface receptors, such as CD4, CCR5, and CXCR4 of T cells and macrophages, are essential for membrane fusion of HIV-1, an initiate step in viral entry. However, it is not well defined how HIV-1 infects CD4-negative mucosal epithelial cells. Here we show that there is a specific interaction between HIV-1 and an invasive oral bacterium, Porphyromonas gingivalis. We found that HIV-1 was trapped on the bacterial surface, which led to internalization of HIV-1 virions as the bacteria invaded CD4-negative epithelial cells. Both bacterial and viral DNA was detected in HeLa and TERT-2 cells exposed to the HIV-1-P. gingivalis complexes 2 hr after the initial infection but not in cells exposed to HIV-1 alone. Moreover, epithelial cell entry of HIV-1 was positively correlated with invasive activity of the P. gingivalis strains tested, even when the binding affinities of HIV-1 to these strains were similar. Finally, it was demonstrated that the viral DNA was integrated into the genome of the host epithelial cells. These results reveal a receptor-independent HIV-1 entry into epithelial cells, which may be relevant in HIV transmission in other mucosal epithelia where complex microbial communities can be found.
Keywords: CCR5; CXCR4; HeLA cells; P. gingivalis 33277; TERT-2 cells; bacterial invasion.
© International & American Associations for Dental Research.
Conflict of interest statement
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Figures



References
-
- Alfsen A, Bomsel M. (2002). HIV-1 gp41 envelope residues 650-685 exposed on native virus act as a lectin to bind epithelial cell galactosyl ceramide. J Biol Chem 277:25649-25659. - PubMed
-
- Bomsel M. (1997). Transcytosis of infectious human immunodeficiency virus across a tight human epithelial cell line barrier. Nat Med 3:42-47. - PubMed
Publication types
MeSH terms
Grants and funding
- U54 CA163069/CA/NCI NIH HHS/United States
- S10 RR025497/RR/NCRR NIH HHS/United States
- U54 CA163072/CA/NCI NIH HHS/United States
- U54 CA163066/CA/NCI NIH HHS/United States
- DE022428/DE/NIDCR NIH HHS/United States
- R21 DE022428/DE/NIDCR NIH HHS/United States
- U54 MD007593/MD/NIMHD NIH HHS/United States
- MD007593/MD/NIMHD NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- DE020915/DE/NIDCR NIH HHS/United States
- UL1 RR024975-01/RR/NCRR NIH HHS/United States
- U54 RR026140/RR/NCRR NIH HHS/United States
- CA163069/CA/NCI NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- G12 MD007586/MD/NIMHD NIH HHS/United States
- RR025497/RR/NCRR NIH HHS/United States
- MD007586/MD/NIMHD NIH HHS/United States
- 2 UL1 TR000445-06/TR/NCATS NIH HHS/United States
- R21 DE020915/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

