PAX6 regulates melanogenesis in the retinal pigmented epithelium through feed-forward regulatory interactions with MITF
- PMID: 24875170
- PMCID: PMC4038462
- DOI: 10.1371/journal.pgen.1004360
PAX6 regulates melanogenesis in the retinal pigmented epithelium through feed-forward regulatory interactions with MITF
Abstract
During organogenesis, PAX6 is required for establishment of various progenitor subtypes within the central nervous system, eye and pancreas. PAX6 expression is maintained in a variety of cell types within each organ, although its role in each lineage and how it acquires cell-specific activity remain elusive. Herein, we aimed to determine the roles and the hierarchical organization of the PAX6-dependent gene regulatory network during the differentiation of the retinal pigmented epithelium (RPE). Somatic mutagenesis of Pax6 in the differentiating RPE revealed that PAX6 functions in a feed-forward regulatory loop with MITF during onset of melanogenesis. PAX6 both controls the expression of an RPE isoform of Mitf and synergizes with MITF to activate expression of genes involved in pigment biogenesis. This study exemplifies how one kernel gene pivotal in organ formation accomplishes a lineage-specific role during terminal differentiation of a single lineage.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
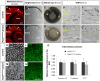
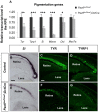
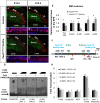
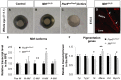

Comment in
-
New functions for old genes: Pax6 and Mitf in eye pigment biogenesis.Pigment Cell Melanoma Res. 2014 Nov;27(6):1005-7. doi: 10.1111/pcmr.12308. Epub 2014 Sep 15. Pigment Cell Melanoma Res. 2014. PMID: 25141763 No abstract available.
Similar articles
-
New functions for old genes: Pax6 and Mitf in eye pigment biogenesis.Pigment Cell Melanoma Res. 2014 Nov;27(6):1005-7. doi: 10.1111/pcmr.12308. Epub 2014 Sep 15. Pigment Cell Melanoma Res. 2014. PMID: 25141763 No abstract available.
-
A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development.PLoS Genet. 2012 Jul;8(7):e1002757. doi: 10.1371/journal.pgen.1002757. Epub 2012 Jul 5. PLoS Genet. 2012. PMID: 22792072 Free PMC article.
-
Retinal pigmented epithelium determination requires the redundant activities of Pax2 and Pax6.Development. 2003 Jul;130(13):2903-15. doi: 10.1242/dev.00450. Development. 2003. PMID: 12756174
-
Pax6: a multi-level regulator of ocular development.Prog Retin Eye Res. 2012 Sep;31(5):351-76. doi: 10.1016/j.preteyeres.2012.04.002. Epub 2012 May 3. Prog Retin Eye Res. 2012. PMID: 22561546 Review.
-
Developmental malformations of the eye: the role of PAX6, SOX2 and OTX2.Clin Genet. 2006 Jun;69(6):459-70. doi: 10.1111/j.1399-0004.2006.00619.x. Clin Genet. 2006. PMID: 16712695 Review.
Cited by
-
New Retinal Pigment Epithelial Cell Model to Unravel Neuroprotection Sensors of Neurodegeneration in Retinal Disease.Front Neurosci. 2022 Jun 30;16:926629. doi: 10.3389/fnins.2022.926629. eCollection 2022. Front Neurosci. 2022. PMID: 35873810 Free PMC article.
-
CHARGE syndrome modeling using patient-iPSCs reveals defective migration of neural crest cells harboring CHD7 mutations.Elife. 2017 Nov 28;6:e21114. doi: 10.7554/eLife.21114. Elife. 2017. PMID: 29179815 Free PMC article.
-
ADAR1-mediated regulation of melanoma invasion.Nat Commun. 2018 May 31;9(1):2154. doi: 10.1038/s41467-018-04600-2. Nat Commun. 2018. PMID: 29855470 Free PMC article.
-
Dynamic enhancer landscapes in human craniofacial development.Nat Commun. 2024 Mar 6;15(1):2030. doi: 10.1038/s41467-024-46396-4. Nat Commun. 2024. PMID: 38448444 Free PMC article.
-
Myelin regulatory factor (MYRF) is a critical early regulator of retinal pigment epithelial development.PLoS Genet. 2025 Apr 15;21(4):e1011670. doi: 10.1371/journal.pgen.1011670. eCollection 2025 Apr. PLoS Genet. 2025. PMID: 40233131 Free PMC article.
References
-
- Strauss O (2005) The retinal pigment epithelium in visual function. Physiol Rev 85: 845–881. - PubMed
-
- del Marmol V, Beermann F (1996) Tyrosinase and related proteins in mammalian pigmentation. FEBS Lett 381: 165–168. - PubMed
-
- Hearing VJ (1999) Biochemical control of melanogenesis and melanosomal organization. The journal of investigative dermatology Symposium proceedings 4: 24–28. - PubMed
-
- Davis N, Yoffe C, Raviv S, Antes R, Berger J, et al. (2009) Pax6 Dosage Requirements in Iris and Ciliary Body Differentiation. Dev Biol 333: 132–42 doi: 10.1016/j.ydbio.2009.06.023 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

