A genome-wide assessment of the role of untagged copy number variants in type 1 diabetes
- PMID: 24875393
- PMCID: PMC4038470
- DOI: 10.1371/journal.pgen.1004367
A genome-wide assessment of the role of untagged copy number variants in type 1 diabetes
Abstract
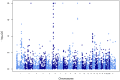
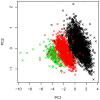
Genome-wide association studies (GWAS) for type 1 diabetes (T1D) have successfully identified more than 40 independent T1D associated tagging single nucleotide polymorphisms (SNPs). However, owing to technical limitations of copy number variants (CNVs) genotyping assays, the assessment of the role of CNVs has been limited to the subset of these in high linkage disequilibrium with tag SNPs. The contribution of untagged CNVs, often multi-allelic and difficult to genotype using existing assays, to the heritability of T1D remains an open question. To investigate this issue, we designed a custom comparative genetic hybridization array (aCGH) specifically designed to assay untagged CNV loci identified from a variety of sources. To overcome the technical limitations of the case control design for this class of CNVs, we genotyped the Type 1 Diabetes Genetics Consortium (T1DGC) family resource (representing 3,903 transmissions from parents to affected offspring) and used an association testing strategy that does not necessitate obtaining discrete genotypes. Our design targeted 4,309 CNVs, of which 3,410 passed stringent quality control filters. As a positive control, the scan confirmed the known T1D association at the INS locus by direct typing of the 5' variable number of tandem repeat (VNTR) locus. Our results clarify the fact that the disease association is indistinguishable from the two main polymorphic allele classes of the INS VNTR, class I-and class III. We also identified novel technical artifacts resulting into spurious associations at the somatically rearranging loci, T cell receptor, TCRA/TCRD and TCRB, and Immunoglobulin heavy chain, IGH, loci on chromosomes 14q11.2, 7q34 and 14q32.33, respectively. However, our data did not identify novel T1D loci. Our results do not support a major role of untagged CNVs in T1D heritability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Rewers M, LaPorte RE, King H, Tuomilehto J (1988) Trends in the prevalence and incidence of diabetes: insulin-dependent diabetes mellitus in childhood. World Health Stat Q 41: : 179–189. Available: http://www.ncbi.nlm.nih.gov/pubmed/2466379. Accessed 11 September 2013. - PubMed
-
- Rewers M (1991) The changing face of the epidemiology of insulin-dependent diabetes mellitus (IDDM): research designs and models of disease causation. Ann Med Ann Med 23: : 419–426 Available: http://www.ncbi.nlm.nih.gov/pubmed/1930939. Accessed 9 August 2013. - PubMed
-
- Bach J-F (2002) The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med 347: : 911–920. Available: http://www.ncbi.nlm.nih.gov/pubmed/12239261. Accessed 6 February 2013. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

