siPools: highly complex but accurately defined siRNA pools eliminate off-target effects
- PMID: 24875475
- PMCID: PMC4081087
- DOI: 10.1093/nar/gku480
siPools: highly complex but accurately defined siRNA pools eliminate off-target effects
Abstract
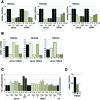
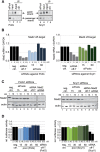
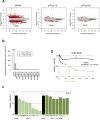
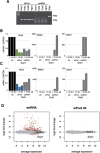
Short interfering RNAs (siRNAs) are widely used as tool for gene inactivation in basic research and therapeutic applications. One of the major shortcomings of siRNA experiments are sequence-specific off-target effects. Such effects are largely unpredictable because siRNAs can affect partially complementary sequences and function like microRNAs (miRNAs), which inhibit gene expression on mRNA stability or translational levels. Here we demonstrate that novel, enzymatically generated siRNA pools-referred to as siPools-containing up to 60 accurately defined siRNAs eliminate off-target effects. This is achieved by the low concentration of each individual siRNA diluting sequence-specific off-target effects below detection limits. In fact, whole transcriptome analyses reveal that single siRNA transfections can severely affect global gene expression. However, when complex siRNA pools are transfected, almost no transcriptome alterations are observed. Taken together, we present enzymatically produced complex but accurately defined siRNA pools with potent on-target silencing but without detectable off-target effects.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Dorsett Y., Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. - PubMed
-
- Meister G., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. - PubMed
-
- Siomi H., Siomi M.C. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. - PubMed
-
- Iwasaki S., Kobayashi M., Yoda M., Sakaguchi Y., Katsuma S., Suzuki T., Tomari Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell. 2010;39:292–299. - PubMed
-
- Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

