Analysis of tetra- and hepta-nucleotides motifs promoting -1 ribosomal frameshifting in Escherichia coli
- PMID: 24875478
- PMCID: PMC4066793
- DOI: 10.1093/nar/gku386
Analysis of tetra- and hepta-nucleotides motifs promoting -1 ribosomal frameshifting in Escherichia coli
Abstract
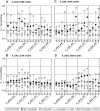
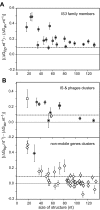
Programmed ribosomal -1 frameshifting is a non-standard decoding process occurring when ribosomes encounter a signal embedded in the mRNA of certain eukaryotic and prokaryotic genes. This signal has a mandatory component, the frameshift motif: it is either a Z_ZZN tetramer or a X_XXZ_ZZN heptamer (where ZZZ and XXX are three identical nucleotides) allowing cognate or near-cognate repairing to the -1 frame of the A site or A and P sites tRNAs. Depending on the signal, the frameshifting frequency can vary over a wide range, from less than 1% to more than 50%. The present study combines experimental and bioinformatics approaches to carry out (i) a systematic analysis of the frameshift propensity of all possible motifs (16 Z_ZZN tetramers and 64 X_XXZ_ZZN heptamers) in Escherichia coli and (ii) the identification of genes potentially using this mode of expression amongst 36 Enterobacteriaceae genomes. While motif efficiency varies widely, a major distinctive rule of bacterial -1 frameshifting is that the most efficient motifs are those allowing cognate re-pairing of the A site tRNA from ZZN to ZZZ. The outcome of the genomic search is a set of 69 gene clusters, 59 of which constitute new candidates for functional utilization of -1 frameshifting.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures







References
-
- Jacks T., Varmus H. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. - PubMed
-
- Jacks T., Power M., Masiarz F., Luciw P., Barr P., Varmus H. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988b;331:280–283. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

