NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism
- PMID: 24875858
- PMCID: PMC4125436
- DOI: 10.1158/2159-8290.CD-14-0014
NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism
Abstract
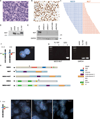
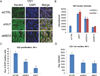
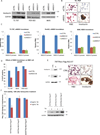
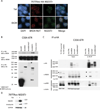
NUT midline carcinoma (NMC) is an aggressive subtype of squamous cell carcinoma that typically harbors BRD4/3-NUT fusion oncoproteins that block differentiation and maintain tumor growth. In 20% of cases, NUT is fused to uncharacterized non-BRD gene(s). We established a new patient-derived NMC cell line (1221) and demonstrated that it harbors a novel NSD3-NUT fusion oncogene. We find that NSD3-NUT is both necessary and sufficient for the blockade of differentiation and maintenance of proliferation in NMC cells. NSD3-NUT binds to BRD4, and BRD bromodomain inhibitors induce differentiation and arrest proliferation of 1221 cells. We find further that NSD3 is required for the blockade of differentiation in BRD4-NUT-expressing NMCs. These findings identify NSD3 as a novel critical oncogenic component and potential therapeutic target in NMC.
Significance: The existence of a family of fusion oncogenes in squamous cell carcinoma is unprecedented, and should lead to key insights into aberrant differentiation in NMC and possibly other squamous cell carcinomas. The involvement of the NSD3 methyltransferase as a component of the NUT fusion protein oncogenic complex identifies a new potential therapeutic target.
©2014 American Association for Cancer Research.
Conflict of interest statement
Disclosure of potential conflicts of interest: No potential conflicts of interest were disclosed.
Figures



References
-
- French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer research. 2003;63(2):304–307. - PubMed
-
- French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27(15):2237–2242. Epub 2007/10/16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

