Rapid evolution of PARP genes suggests a broad role for ADP-ribosylation in host-virus conflicts
- PMID: 24875882
- PMCID: PMC4038475
- DOI: 10.1371/journal.pgen.1004403
Rapid evolution of PARP genes suggests a broad role for ADP-ribosylation in host-virus conflicts
Abstract
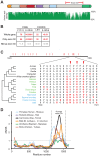
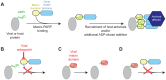
Post-translational protein modifications such as phosphorylation and ubiquitinylation are common molecular targets of conflict between viruses and their hosts. However, the role of other post-translational modifications, such as ADP-ribosylation, in host-virus interactions is less well characterized. ADP-ribosylation is carried out by proteins encoded by the PARP (also called ARTD) gene family. The majority of the 17 human PARP genes are poorly characterized. However, one PARP protein, PARP13/ZAP, has broad antiviral activity and has evolved under positive (diversifying) selection in primates. Such evolution is typical of domains that are locked in antagonistic 'arms races' with viral factors. To identify additional PARP genes that may be involved in host-virus interactions, we performed evolutionary analyses on all primate PARP genes to search for signatures of rapid evolution. Contrary to expectations that most PARP genes are involved in 'housekeeping' functions, we found that nearly one-third of PARP genes are evolving under strong recurrent positive selection. We identified a >300 amino acid disordered region of PARP4, a component of cytoplasmic vault structures, to be rapidly evolving in several mammalian lineages, suggesting this region serves as an important host-pathogen specificity interface. We also found positive selection of PARP9, 14 and 15, the only three human genes that contain both PARP domains and macrodomains. Macrodomains uniquely recognize, and in some cases can reverse, protein mono-ADP-ribosylation, and we observed strong signatures of recurrent positive selection throughout the macro-PARP macrodomains. Furthermore, PARP14 and PARP15 have undergone repeated rounds of gene birth and loss during vertebrate evolution, consistent with recurrent gene innovation. Together with previous studies that implicated several PARPs in immunity, as well as those that demonstrated a role for virally encoded macrodomains in host immune evasion, our evolutionary analyses suggest that addition, recognition and removal of ADP-ribosylation is a critical, underappreciated currency in host-virus conflicts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Adhya D, Basu A (2010) Epigenetic modulation of host: new insights into immune evasion by viruses. J Biosci 35: 647–663. - PubMed
-
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F (2010) Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci 35: 208–219. - PubMed
-
- Ziegler M (2000) New functions of a long-known molecule. Emerging roles of NAD in cellular signaling. Eur J Biochem 267: 1550–1564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

