The Fab conformations in the solution structure of human immunoglobulin G4 (IgG4) restrict access to its Fc region: implications for functional activity
- PMID: 24876381
- PMCID: PMC4110284
- DOI: 10.1074/jbc.M114.572404
The Fab conformations in the solution structure of human immunoglobulin G4 (IgG4) restrict access to its Fc region: implications for functional activity
Abstract
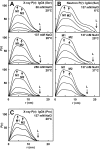
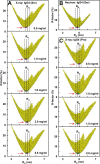
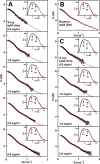
Human IgG4 antibody shows therapeutically useful properties compared with the IgG1, IgG2, and IgG3 subclasses. Thus IgG4 does not activate complement and shows conformational variability. These properties are attributable to its hinge region, which is the shortest of the four IgG subclasses. Using high throughput scattering methods, we studied the solution structure of wild-type IgG4(Ser(222)) and a hinge mutant IgG4(Pro(222)) in different buffers and temperatures where the proline substitution suppresses the formation of half-antibody. Analytical ultracentrifugation showed that both IgG4 forms were principally monomeric with sedimentation coefficients s20,w(0) of 6.6-6.8 S. A monomer-dimer equilibrium was observed in heavy water buffer at low temperature. Scattering showed that the x-ray radius of gyration Rg was unchanged with concentration in 50-250 mm NaCl buffers, whereas the neutron Rg values showed a concentration-dependent increase as the temperature decreased in heavy water buffers. The distance distribution curves (P(r)) revealed two peaks, M1 and M2, that shifted below 2 mg/ml to indicate concentration-dependent IgG4 structures in addition to IgG4 dimer formation at high concentration in heavy water. Constrained x-ray and neutron scattering modeling revealed asymmetric solution structures for IgG4(Ser(222)) with extended hinge structures. The IgG4(Pro(222)) structure was similar. Both IgG4 structures showed that their Fab regions were positioned close enough to the Fc region to restrict C1q binding. Our new molecular models for IgG4 explain its inability to activate complement and clarify aspects of its stability and function for therapeutic applications.
Keywords: Analytical Ultracentrifugation; Antibody; Complement; Constrained Modeling; Human IgG4; Neutron Scattering; Protein Structure; X-ray Scattering.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Hamilton R. G. (2001) The Human IgG Subclasses, Calbiochem, La Jolla, CA
-
- Aalberse R. C., Schuurman J., van Ree R. (1999) The apparent monovalency of human IgG4 is due to bispecificity. Int. Arch. Allergy Immunol. 118, 187–189 - PubMed
-
- Rispens T., Ooievaar-De Heer P., Vermeulen E., Schuurman J., van der Neut Kolfschoten M., Aalberse R. C. (2009) Human IgG4 binds to IgG4 and conformationally altered IgG1 via Fc-Fc interactions. J. Immunol. 182, 4275–4281 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

