Synthetic zinc finger proteins: the advent of targeted gene regulation and genome modification technologies
- PMID: 24877793
- PMCID: PMC4139171
- DOI: 10.1021/ar500039w
Synthetic zinc finger proteins: the advent of targeted gene regulation and genome modification technologies
Abstract
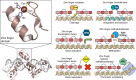
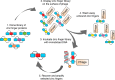
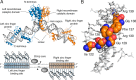
The understanding of gene regulation and the structure and function of the human genome increased dramatically at the end of the 20th century. Yet the technologies for manipulating the genome have been slower to develop. For instance, the field of gene therapy has been focused on correcting genetic diseases and augmenting tissue repair for more than 40 years. However, with the exception of a few very low efficiency approaches, conventional genetic engineering methods have only been able to add auxiliary genes to cells. This has been a substantial obstacle to the clinical success of gene therapies and has also led to severe unintended consequences in several cases. Therefore, technologies that facilitate the precise modification of cellular genomes have diverse and significant implications in many facets of research and are essential for translating the products of the Genomic Revolution into tangible benefits for medicine and biotechnology. To address this need, in the 1990s, we embarked on a mission to develop technologies for engineering protein-DNA interactions with the aim of creating custom tools capable of targeting any DNA sequence. Our goal has been to allow researchers to reach into genomes to specifically regulate, knock out, or replace any gene. To realize these goals, we initially focused on understanding and manipulating zinc finger proteins. In particular, we sought to create a simple and straightforward method that enables unspecialized laboratories to engineer custom DNA-modifying proteins using only defined modular components, a web-based utility, and standard recombinant DNA technology. Two significant challenges we faced were (i) the development of zinc finger domains that target sequences not recognized by naturally occurring zinc finger proteins and (ii) determining how individual zinc finger domains could be tethered together as polydactyl proteins to recognize unique locations within complex genomes. We and others have since used this modular assembly method to engineer artificial proteins and enzymes that activate, repress, or create defined changes to user-specified genes in human cells, plants, and other organisms. We have also engineered novel methods for externally controlling protein activity and delivery, as well as developed new strategies for the directed evolution of protein and enzyme function. This Account summarizes our work in these areas and highlights independent studies that have successfully used the modular assembly approach to create proteins with novel function. We also discuss emerging alternative methods for genomic targeting, including transcription activator-like effectors (TALEs) and CRISPR/Cas systems, and how they complement the synthetic zinc finger protein technology.
Figures
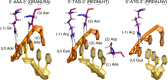

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

