GATA4 represses an ileal program of gene expression in the proximal small intestine by inhibiting the acetylation of histone H3, lysine 27
- PMID: 24878542
- PMCID: PMC4684644
- DOI: 10.1016/j.bbagrm.2014.05.018
GATA4 represses an ileal program of gene expression in the proximal small intestine by inhibiting the acetylation of histone H3, lysine 27
Abstract
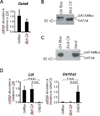
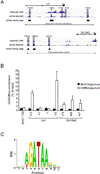
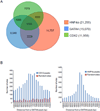
GATA4 is expressed in the proximal 85% of small intestine where it promotes a proximal intestinal ('jejunal') identity while repressing a distal intestinal ('ileal') identity, but its molecular mechanisms are unclear. Here, we tested the hypothesis that GATA4 promotes a jejunal versus ileal identity in mouse intestine by directly activating and repressing specific subsets of absorptive enterocyte genes by modulating the acetylation of histone H3, lysine 27 (H3K27), a mark of active chromatin, at sites of GATA4 occupancy. Global analysis of mouse jejunal epithelium showed a statistically significant association of GATA4 occupancy with GATA4-regulated genes. Occupancy was equally distributed between down- and up-regulated targets, and occupancy sites showed a dichotomy of unique motif over-representation at down- versus up-regulated genes. H3K27ac enrichment at GATA4-binding loci that mapped to down-regulated genes (activation targets) was elevated, changed little upon conditional Gata4 deletion, and was similar to control ileum, whereas H3K27ac enrichment at GATA4-binding loci that mapped to up-regulated genes (repression targets) was depleted, increased upon conditional Gata4 deletion, and approached H3K27ac enrichment in wild-type control ileum. These data support the hypothesis that GATA4 both activates and represses intestinal genes, and show that GATA4 represses an ileal program of gene expression in the proximal small intestine by inhibiting the acetylation of H3K27.
Keywords: Chromatin occupancy; GATA4; H3K27ac; Histone modification; Intestinal epithelium; Transcriptional repression.
Copyright © 2014 Elsevier B.V. All rights reserved.
Figures





References
-
- van Wering HM, Bosse T, Musters A, de Jong E, de Jong N, Hogen Esch CE, Boudreau F, Swain GP, Dowling LN, Montgomery RK, et al. Complex regulation of the lactase-phlorizin hydrolase promoter by GATA-4. Am J Physio Gastrointest Liver Physiol. 2004;287(4):G899–G909. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

