TbKAP6, a mitochondrial HMG box-containing protein in Trypanosoma brucei, is the first trypanosomatid kinetoplast-associated protein essential for kinetoplast DNA replication and maintenance
- PMID: 24879122
- PMCID: PMC4135736
- DOI: 10.1128/EC.00260-13
TbKAP6, a mitochondrial HMG box-containing protein in Trypanosoma brucei, is the first trypanosomatid kinetoplast-associated protein essential for kinetoplast DNA replication and maintenance
Abstract
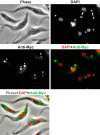
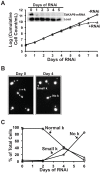
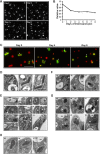
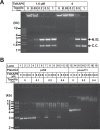
Kinetoplast DNA (kDNA), the mitochondrial genome of trypanosomatids, is a giant planar network of catenated minicircles and maxicircles. In vivo kDNA is organized as a highly condensed nucleoprotein disk. So far, in Trypanosoma brucei, proteins involved in the maintenance of the kDNA condensed structure remain poorly characterized. In Crithidia fasciculata, some small basic histone H1-like kinetoplast-associated proteins (CfKAP) have been shown to condense isolated kDNA networks in vitro. High-mobility group (HMG) box-containing proteins, such as mitochondrial transcription factor A (TFAM) in mammalian cells and Abf2 in the budding yeast, have been shown essential for the packaging of mitochondrial DNA (mtDNA) into mitochondrial nucleoids, remodeling of mitochondrial nucleoids, gene expression, and maintenance of mtDNA. Here, we report that TbKAP6, a mitochondrial HMG box-containing protein, is essential for parasite cell viability and involved in kDNA replication and maintenance. The RNA interference (RNAi) depletion of TbKAP6 stopped cell growth. Replication of both minicircles and maxicircles was inhibited. RNAi or overexpression of TbKAP6 resulted in the disorganization, shrinkage, and loss of kDNA. Minicircle release, the first step in kDNA replication, was inhibited immediately after induction of RNAi, but it quickly increased 3-fold upon overexpression of TbKAP6. Since the release of covalently closed minicircles is mediated by a type II topoisomerase (topo II), we examined the potential interactions between TbKAP6 and topo II. Recombinant TbKAP6 (rTbKAP6) promotes the topo II-mediated decatenation of kDNA. rTbKAP6 can condense isolated kDNA networks in vitro. These results indicate that TbKAP6 is involved in the replication and maintenance of kDNA.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
TbPIF1, a Trypanosoma brucei mitochondrial DNA helicase, is essential for kinetoplast minicircle replication.J Biol Chem. 2010 Mar 5;285(10):7056-66. doi: 10.1074/jbc.M109.084038. Epub 2009 Dec 30. J Biol Chem. 2010. PMID: 20042610 Free PMC article.
-
Trypanosoma brucei Tb927.2.6100 is an essential protein associated with kinetoplast DNA.Eukaryot Cell. 2013 Jul;12(7):970-8. doi: 10.1128/EC.00352-12. Epub 2013 May 6. Eukaryot Cell. 2013. PMID: 23650088 Free PMC article.
-
Stem-loop silencing reveals that a third mitochondrial DNA polymerase, POLID, is required for kinetoplast DNA replication in trypanosomes.Eukaryot Cell. 2008 Dec;7(12):2141-6. doi: 10.1128/EC.00199-08. Epub 2008 Oct 10. Eukaryot Cell. 2008. PMID: 18849470 Free PMC article.
-
The structure and replication of kinetoplast DNA.Curr Mol Med. 2004 Sep;4(6):623-47. doi: 10.2174/1566524043360096. Curr Mol Med. 2004. PMID: 15357213 Review.
-
The structure and replication of kinetoplast DNA.Annu Rev Microbiol. 1995;49:117-43. doi: 10.1146/annurev.mi.49.100195.001001. Annu Rev Microbiol. 1995. PMID: 8561456 Review.
Cited by
-
Mitochondrial genome maintenance-the kinetoplast story.FEMS Microbiol Rev. 2023 Nov 1;47(6):fuac047. doi: 10.1093/femsre/fuac047. FEMS Microbiol Rev. 2023. PMID: 36449697 Free PMC article. Review.
-
UMSBP2 is chromatin remodeler that functions in regulation of gene expression and suppression of antigenic variation in trypanosomes.Nucleic Acids Res. 2023 Jun 23;51(11):5678-5698. doi: 10.1093/nar/gkad402. Nucleic Acids Res. 2023. PMID: 37207337 Free PMC article.
-
Crystal structure of the HMGA AT-hook 1 domain bound to the minor groove of AT-rich DNA and inhibition by antikinetoplastid drugs.Sci Rep. 2024 Oct 30;14(1):26173. doi: 10.1038/s41598-024-77522-3. Sci Rep. 2024. PMID: 39478017 Free PMC article.
-
Characterization of a novel Leishmania antigen containing a repetitive domain and its potential use as a prophylactic and therapeutic vaccine.mSphere. 2025 May 27;10(5):e0009725. doi: 10.1128/msphere.00097-25. Epub 2025 Apr 22. mSphere. 2025. PMID: 40261025 Free PMC article.
-
Oxidative Phosphorylation Is Required for Powering Motility and Development of the Sleeping Sickness Parasite Trypanosoma brucei in the Tsetse Fly Vector.mBio. 2022 Feb 22;13(1):e0235721. doi: 10.1128/mbio.02357-21. Epub 2022 Jan 11. mBio. 2022. PMID: 35012336 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

