ATG13: just a companion, or an executor of the autophagic program?
- PMID: 24879146
- PMCID: PMC4091178
- DOI: 10.4161/auto.28987
ATG13: just a companion, or an executor of the autophagic program?
Erratum in
- Autophagy. 2014 Aug;10(8):1481
Abstract
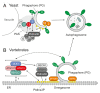
During the past 20 years, autophagy signaling has entered the main stage of the cell biological theater. Autophagy represents an intracellular degradation process that is involved in both the bulk recycling of cytoplasmic components and the selective removal of organelles, protein aggregates, or intracellular pathogens. The understanding of autophagy has been greatly facilitated by the characterization of the molecular machinery governing this process. In yeast, initiation of autophagy is controlled by the Atg1 kinase complex, which is composed of the Ser/Thr kinase Atg1, the adaptor protein Atg13, and the ternary complex of Atg17-Atg31-Atg29. In vertebrates, the orthologous ULK1 kinase complex contains the Ser/Thr kinase ULK1 and the accessory proteins ATG13, RB1CC1, and ATG101. Among these components, Atg1/ULK1 have gained major attention in the past, i.e., for the identification of upstream regulatory kinases, the characterization of downstream substrates controlling the autophagic flux, or as a druggable target for the modulation of autophagy. However, accumulating data indicate that the function of Atg13/ATG13 has been likely underestimated so far. In addition to ensuring proper Atg1/ULK1 recruitment and activity, this adaptor molecule has been implicated in ULK1-independent autophagy processes. Furthermore, recent data have identified additional binding partners of Atg13/ATG13 besides the components of the Atg1/ULK1 complex, e.g., Atg8 family proteins or acidic phospholipids. Therefore, in this review we will center the spotlight on Atg13/ATG13 and summarize the role that Atg13/ATG13 assumes in the autophagy stage play.
Keywords: ATG101; ATG13; RB1CC1; ULK1; autophagy.
Figures
References
-
- Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy. 2008;4:740–3. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
