A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans
- PMID: 24879462
- PMCID: PMC4125384
- DOI: 10.1534/genetics.114.166389
A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans
Abstract
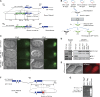
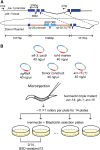
Genome editing based on CRISPR (clustered regularly interspaced short palindromic repeats)-associated nuclease (Cas9) has been successfully applied in dozens of diverse plant and animal species, including the nematode Caenorhabditis elegans. The rapid life cycle and easy access to the ovary by micro-injection make C. elegans an ideal organism both for applying CRISPR-Cas9 genome editing technology and for optimizing genome-editing protocols. Here we report efficient and straightforward CRISPR-Cas9 genome-editing methods for C. elegans, including a Co-CRISPR strategy that facilitates detection of genome-editing events. We describe methods for detecting homologous recombination (HR) events, including direct screening methods as well as new selection/counterselection strategies. Our findings reveal a surprisingly high frequency of HR-mediated gene conversion, making it possible to rapidly and precisely edit the C. elegans genome both with and without the use of co-inserted marker genes.
Keywords: CRISPR-Cas9 system; CRISPR-Cas9-induced indels; CRISPR-Cas9-mediated HR; Co-CRISPR; blasticidin selection.
Copyright © 2014 by the Genetics Society of America.
Figures


References
-
- Bhaya D., Davison M., Barrangou R., 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45: 273–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

