Epigenetic regulation of fetal globin gene expression in adult erythroid cells
- PMID: 24880147
- PMCID: PMC4227965
- DOI: 10.1016/j.trsl.2014.05.002
Epigenetic regulation of fetal globin gene expression in adult erythroid cells
Abstract
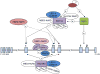
The developmental regulation of globin gene expression has served as an important model for understanding higher eukaryotic transcriptional control mechanisms. During human erythroid development, there is a sequential switch from expression of the embryonic ε-globin gene to the fetal ɣ-globin gene in utero, and postpartum the ɣ-globin gene is silenced, as the β-globin gene becomes the predominantly expressed locus. Because the expression of normally silenced fetal ɣ-type globin genes and resultant production of fetal hemoglobin (HbF) in adult erythroid cells can ameliorate the pathophysiological consequences of both abnormal β-globin chains in sickle cell anemia and deficient β-globin chain production in β-thalassemia, understanding the complex mechanisms of this developmental switch has direct translational clinical relevance. Of particular interest for translational research are the factors that mediate silencing of the ɣ-globin gene in adult stage erythroid cells. In addition to the regulatory roles of transcription factors and their cognate DNA sequence motifs, there has been a growing appreciation of the role of epigenetic signals and their cognate factors in gene regulation, and in particular in gene silencing through chromatin. Much of the information about epigenetic silencing stems from studies of globin gene regulation. As discussed here, the term epigenetics refers to postsynthetic modifications of DNA and chromosomal histone proteins that affect gene expression and can be inherited through somatic cell replication. A full understanding of the molecular mechanisms of epigenetic silencing of HbF expression should facilitate the development of more effective treatment of β-globin chain hemoglobinopathies.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
-
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–32. - PubMed
-
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. - PubMed
-
- McGhee JD, Ginder GD. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–20. - PubMed
-
- Razin A, Riggs AD. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

