A SET-domain-independent role of WRAD complex in cell-cycle regulatory function of mixed lineage leukemia
- PMID: 24880690
- PMCID: PMC4081079
- DOI: 10.1093/nar/gku458
A SET-domain-independent role of WRAD complex in cell-cycle regulatory function of mixed lineage leukemia
Abstract
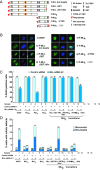
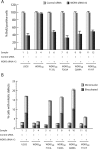
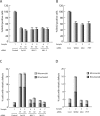
MLL, the trithorax ortholog, is a well-characterized histone 3 lysine 4 methyltransferase that is crucial for proper regulation of the Hox genes during embryonic development. Chromosomal translocations, disrupting the Mll gene, lead to aggressive leukemia with poor prognosis. However, the functions of MLL in cellular processes like cell-cycle regulation are not well studied. Here we show that the MLL has a regulatory role during multiple phases of the cell cycle. RNAi-mediated knockdown reveals that MLL regulates S-phase progression and, proper segregation and cytokinesis during M phase. Using deletions and mutations, we narrow the cell-cycle regulatory role to the C subunit of MLL. Our analysis reveals that the transactivation domain and not the SET domain is important for the S-phase function of MLL. Surprisingly, disruption of MLL-WRAD interaction is sufficient to disrupt proper mitotic progression. These mitotic functions of WRAD are independent of SET domain of MLL and, therefore, define a new role of WRAD in subset of MLL functions. Finally, we address the overlapping and unique roles of the different SET family members in the cell cycle.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

