Studying tumorigenesis through network evolution and somatic mutational perturbations in the cancer interactome
- PMID: 24881052
- PMCID: PMC4104318
- DOI: 10.1093/molbev/msu167
Studying tumorigenesis through network evolution and somatic mutational perturbations in the cancer interactome
Abstract
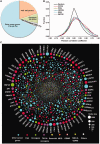
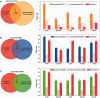
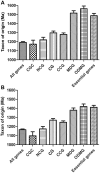
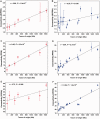
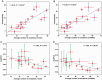
Cells govern biological functions through complex biological networks. Perturbations to networks may drive cells to new phenotypic states, for example, tumorigenesis. Identifying how genetic lesions perturb molecular networks is a fundamental challenge. This study used large-scale human interactome data to systematically explore the relationship among network topology, somatic mutation, evolutionary rate, and evolutionary origin of cancer genes. We found the unique network centrality of cancer proteins, which is largely independent of gene essentiality. Cancer genes likely have experienced a lower evolutionary rate and stronger purifying selection than those of noncancer, Mendelian disease, and orphan disease genes. Cancer proteins tend to have ancient histories, likely originated in early metazoan, although they are younger than proteins encoded by Mendelian disease genes, orphan disease genes, and essential genes. We found that the protein evolutionary origin (age) positively correlates with protein connectivity in the human interactome. Furthermore, we investigated the network-attacking perturbations due to somatic mutations identified from 3,268 tumors across 12 cancer types in The Cancer Genome Atlas. We observed a positive correlation between protein connectivity and the number of nonsynonymous somatic mutations, whereas a weaker or insignificant correlation between protein connectivity and the number of synonymous somatic mutations. These observations suggest that somatic mutational network-attacking perturbations to hub genes play an important role in tumor emergence and evolution. Collectively, this work has broad biomedical implications for both basic cancer biology and the development of personalized cancer therapy.
Keywords: TCGA; network evolution; network-attacking perturbation; somatic mutation; tumorigenesis.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Aparicio S, Caldas C. The implications of clonal genome evolution for cancer medicine. N Engl J Med. 2013;368:842–851. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

