Bioattractors: dynamical systems theory and the evolution of regulatory processes
- PMID: 24882812
- PMCID: PMC4048087
- DOI: 10.1113/jphysiol.2014.272385
Bioattractors: dynamical systems theory and the evolution of regulatory processes
Abstract
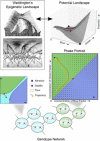
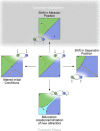
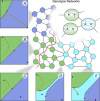
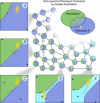
In this paper, we illustrate how dynamical systems theory can provide a unifying conceptual framework for evolution of biological regulatory systems. Our argument is that the genotype-phenotype map can be characterized by the phase portrait of the underlying regulatory process. The features of this portrait--such as attractors with associated basins and their bifurcations--define the regulatory and evolutionary potential of a system. We show how the geometric analysis of phase space connects Waddington's epigenetic landscape to recent computational approaches for the study of robustness and evolvability in network evolution. We discuss how the geometry of phase space determines the probability of possible phenotypic transitions. Finally, we demonstrate how the active, self-organizing role of the environment in phenotypic evolution can be understood in terms of dynamical systems concepts. This approach yields mechanistic explanations that go beyond insights based on the simulation of evolving regulatory networks alone. Its predictions can now be tested by studying specific, experimentally tractable regulatory systems using the tools of modern systems biology. A systematic exploration of such systems will enable us to understand better the nature and origin of the phenotypic variability, which provides the substrate for evolution by natural selection.
© 2014 The Authors. The Journal of Physiology © 2014 The Physiological Society.
Figures

References
-
- Alberch P. Developmental constraints in evolutionary processes. In: Bonner JT, editor. Evolution and Development. Heidelberg: Springer; 1982. pp. 313–332.
-
- Alberch P. From genes to phenotype: dynamical systems and evolvability. Genetica. 1991;84:5–11. - PubMed
-
- Arthur W. Biased Embryos and Evolution. Cambridge: Cambridge University Press; 2004.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

