Identification of a 20-gene expression-based risk score as a predictor of clinical outcome in chronic lymphocytic leukemia patients
- PMID: 24883311
- PMCID: PMC4026849
- DOI: 10.1155/2014/423174
Identification of a 20-gene expression-based risk score as a predictor of clinical outcome in chronic lymphocytic leukemia patients
Abstract
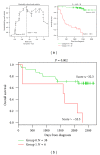
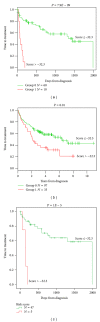
Despite the improvement in treatment options, chronic lymphocytic leukemia (CLL) remains an incurable disease and patients show a heterogeneous clinical course requiring therapy for many of them. In the current work, we have built a 20-gene expression (GE)-based risk score predictive for patients overall survival and improving risk classification using microarray gene expression data. GE-based risk score allowed identifying a high-risk group associated with a significant shorter overall survival (OS) and time to treatment (TTT) (P ≤ .01), comprising 19.6% and 13.6% of the patients in two independent cohorts. GE-based risk score, and NRIP1 and TCF7 gene expression remained independent prognostic factors using multivariate Cox analyses and combination of GE-based risk score together with NRIP1 and TCF7 gene expression enabled the identification of three clinically distinct groups of CLL patients. Therefore, this GE-based risk score represents a powerful tool for risk stratification and outcome prediction of CLL patients and could thus be used to guide clinical and therapeutic decisions prospectively.
Figures


References
-
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. The New England Journal of Medicine. 2005;352(8):804–815. - PubMed
-
- Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. - PMC - PubMed
-
- Stamatopoulos K, Belessi C, Moreno C, et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: pathogenetic implications and clinical correlations. Blood. 2007;109(1):259–270. - PubMed
-
- Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. The New England Journal of Medicine. 2004;351(9):893–901. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

