Bioactive oligosaccharide natural products
- PMID: 24883430
- PMCID: PMC5267508
- DOI: 10.1039/c3np70128j
Bioactive oligosaccharide natural products
Abstract
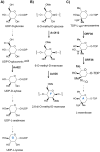
Covering up to December 2013. Oligosaccharide natural products target a wide spectrum of biological processes including disruption of cell wall biosynthesis, interference of bacterial translation, and inhibition of human α-amylase. Correspondingly, oligosaccharides possess the potential for development as treatments of such diverse diseases as bacterial infections and type II diabetes. Despite their potent and selective activities and potential clinical relevance, isolated bioactive secondary metabolic oligosaccharides are less prevalent than other classes of natural products and their biosynthesis has received comparatively less attention. This review highlights the unique modes of action and biosynthesis of four classes of bioactive oligosaccharides: the orthosomycins, moenomycins, saccharomicins, and acarviostatins.
Figures













Similar articles
-
Microbial Oligosaccharides with Biomedical Applications.Mar Drugs. 2021 Jun 21;19(6):350. doi: 10.3390/md19060350. Mar Drugs. 2021. PMID: 34205503 Free PMC article. Review.
-
Glycosyltransferases involved in the biosynthesis of biologically active natural products that contain oligosaccharides.Mol Biosyst. 2005 Jul;1(2):117-26. doi: 10.1039/b503215f. Epub 2005 Jun 23. Mol Biosyst. 2005. PMID: 16880973 Review.
-
Marine actinomycetes-derived angucyclines and angucyclinones with biosynthesis and activity--past 10 years (2014-2023).Eur J Med Chem. 2025 Feb 5;283:117161. doi: 10.1016/j.ejmech.2024.117161. Epub 2024 Dec 11. Eur J Med Chem. 2025. PMID: 39700875 Review.
-
Bioactive Pimarane-Type Diterpenes from Marine Organisms.Chem Biodivers. 2018 Jan;15(1). doi: 10.1002/cbdv.201700276. Epub 2017 Dec 21. Chem Biodivers. 2018. PMID: 28737020 Review.
-
Four acarviosin-containing oligosaccharides identified from Streptomyces coelicoflavus ZG0656 are potent inhibitors of alpha-amylase.Carbohydr Res. 2008 Apr 7;343(5):882-92. doi: 10.1016/j.carres.2008.01.020. Epub 2008 Jan 26. Carbohydr Res. 2008. PMID: 18294624
Cited by
-
Novofumigatonin biosynthesis involves a non-heme iron-dependent endoperoxide isomerase for orthoester formation.Nat Commun. 2018 Jul 3;9(1):2587. doi: 10.1038/s41467-018-04983-2. Nat Commun. 2018. PMID: 29968715 Free PMC article.
-
A standardized workflow for submitting data to the Minimum Information about a Biosynthetic Gene cluster (MIBiG) repository: prospects for research-based educational experiences.Stand Genomic Sci. 2018 Jul 11;13:16. doi: 10.1186/s40793-018-0318-y. eCollection 2018. Stand Genomic Sci. 2018. PMID: 30008988 Free PMC article.
-
Rapid Mining of Novel α-Glucosidase and Lipase Inhibitors from Streptomyces sp. HO1518 Using UPLC-QTOF-MS/MS.Mar Drugs. 2022 Mar 4;20(3):189. doi: 10.3390/md20030189. Mar Drugs. 2022. PMID: 35323488 Free PMC article.
-
Naturally Occurring Organohalogen Compounds-A Comprehensive Review.Prog Chem Org Nat Prod. 2023;121:1-546. doi: 10.1007/978-3-031-26629-4_1. Prog Chem Org Nat Prod. 2023. PMID: 37488466 Review.
-
Enterococcus faecalis YM0831 suppresses sucrose-induced hyperglycemia in a silkworm model and in humans.Commun Biol. 2019 May 2;2:157. doi: 10.1038/s42003-019-0407-5. eCollection 2019. Commun Biol. 2019. PMID: 31069266 Free PMC article. Clinical Trial.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

