Molecular evolution and functional characterisation of an ancient phenylalanine ammonia-lyase gene (NnPAL1) from Nelumbo nucifera: novel insight into the evolution of the PAL family in angiosperms
- PMID: 24884360
- PMCID: PMC4102242
- DOI: 10.1186/1471-2148-14-100
Molecular evolution and functional characterisation of an ancient phenylalanine ammonia-lyase gene (NnPAL1) from Nelumbo nucifera: novel insight into the evolution of the PAL family in angiosperms
Abstract
Background: Phenylalanine ammonia-lyase (PAL; E.C.4.3.1.5) is a key enzyme of the phenylpropanoid pathway in plant development, and it catalyses the deamination of phenylalanine to trans-cinnamic acid, leading to the production of secondary metabolites. This enzyme has been identified in many organisms, ranging from prokaryotes to higher plants. Because Nelumbo nucifera is a basal dicot rich in many secondary metabolites, it is a suitable candidate for research on the phenylpropanoid pathway.
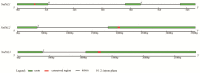
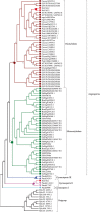
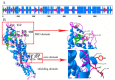
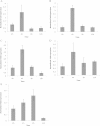
Results: Three PAL members, NnPAL1, NnPAL2 and NnPAL3, have been identified in N. nucifera using genome-wide analysis. NnPAL1 contains two introns; however, both NnPAL2 and NnPAL3 have only one intron. Molecular and evolutionary analysis of NnPAL1 confirms that it is an ancient PAL member of the angiosperms and may have a different origin. However, PAL clusters, except NnPAL1, are monophyletic after the split between dicots and monocots. These observations suggest that duplication events remain an important occurrence in the evolution of the PAL gene family. Molecular assays demonstrate that the mRNA of the NnPAL1 gene is 2343 bp in size and encodes a 717 amino acid polypeptide. The optimal pH and temperature of the recombinant NnPAL1 protein are 9.0 and 55°C, respectively. The NnPAL1 protein retains both PAL and weak TAL catalytic activities with Km values of 1.07 mM for L-phenylalanine and 3.43 mM for L-tyrosine, respectively. Cis-elements response to environmental stress are identified and confirmed using real-time PCR for treatments with abscisic acid (ABA), indoleacetic acid (IAA), ultraviolet light, Neurospora crassa (fungi) and drought.
Conclusions: We conclude that the angiosperm PAL genes are not derived from a single gene in an ancestral angiosperm genome; therefore, there may be another ancestral duplication and vertical inheritance from the gymnosperms. The different evolutionary histories for PAL genes in angiosperms suggest different mechanisms of functional regulation. The expression patterns of NnPAL1 in response to stress may be necessary for the survival of N. nucifera since the Cretaceous Period. The discovery and characterisation of the ancient NnPAL1 help to elucidate PAL evolution in angiosperms.
Figures

Similar articles
-
The phenylalanine ammonia lyase (PAL) gene family shows a gymnosperm-specific lineage.BMC Genomics. 2012 Jun 11;13 Suppl 3(Suppl 3):S1. doi: 10.1186/1471-2164-13-S3-S1. BMC Genomics. 2012. PMID: 22759610 Free PMC article.
-
Evolution of phenylalanine ammonia-lyase protein family from algae to angiosperm.Funct Integr Genomics. 2025 Feb 18;25(1):40. doi: 10.1007/s10142-025-01548-7. Funct Integr Genomics. 2025. PMID: 39966266
-
Genome-wide identification of the phenylalanine ammonia-lyase gene from Epimedium Pubescens Maxim. (Berberidaceae): novel insight into the evolution of the PAL gene family.BMC Plant Biol. 2024 Sep 4;24(1):831. doi: 10.1186/s12870-024-05480-z. BMC Plant Biol. 2024. PMID: 39232677 Free PMC article.
-
A modern view of phenylalanine ammonia lyase.Biochem Cell Biol. 2007 Jun;85(3):273-82. doi: 10.1139/o07-018. Biochem Cell Biol. 2007. PMID: 17612622 Review.
-
Biotechnological production and applications of microbial phenylalanine ammonia lyase: a recent review.Crit Rev Biotechnol. 2014 Sep;34(3):258-68. doi: 10.3109/07388551.2013.791660. Epub 2013 May 21. Crit Rev Biotechnol. 2014. PMID: 23688066 Review.
Cited by
-
The first step into phenolic metabolism in the hornwort Anthoceros agrestis: molecular and biochemical characterization of two phenylalanine ammonia-lyase isoforms.Planta. 2022 Jul 7;256(2):33. doi: 10.1007/s00425-022-03944-w. Planta. 2022. PMID: 35796843 Free PMC article.
-
Isolation and Functional Characterization of a Phenylalanine Ammonia-Lyase Gene (SsPAL1) from Coleus (Solenostemon scutellarioides (L.) Codd).Molecules. 2015 Sep 16;20(9):16833-51. doi: 10.3390/molecules200916833. Molecules. 2015. PMID: 26389875 Free PMC article.
-
Molecular dynamics simulation and bioinformatics study on chloroplast stromal ridge complex from rice (Oryza sativa L.).BMC Bioinformatics. 2016 Jan 12;17:28. doi: 10.1186/s12859-016-0877-0. BMC Bioinformatics. 2016. PMID: 26753869 Free PMC article.
-
Molecular regulation of immunity in tea plants.Mol Biol Rep. 2023 Mar;50(3):2883-2892. doi: 10.1007/s11033-022-08177-4. Epub 2022 Dec 20. Mol Biol Rep. 2023. PMID: 36538170 Review.
-
Genome-wide identification and expression analysis of phenylalanine ammonia-lyase (PAL) family in rapeseed (Brassica napus L.).BMC Plant Biol. 2023 Oct 10;23(1):481. doi: 10.1186/s12870-023-04472-9. BMC Plant Biol. 2023. PMID: 37814209 Free PMC article.
References
-
- Pellegrini L, Rohfritsch O, Fritig B, Legrand M. Phenylalanine ammonia-lyase in tobacco: molecular cloning and gene expression during the hypersensitive reaction to tobacco mosaic virus and the response to a fungal elicitor. Plant Physiol. 1994;106:877–886. doi: 10.1104/pp.106.3.877. - DOI - PMC - PubMed
-
- Hamberger B, Ellis M, Friedmann M, de Azevedo SC, Barbazuk B, Douglas CJ. Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: the Populus lignin toolbox and conservation and diversification of angiosperm gene families. Can J Bot. 2007;85(12):1182–1201. doi: 10.1139/B07-098. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

