Multilocus sequence typing and ftsI sequencing: a powerful tool for surveillance of penicillin-binding protein 3-mediated beta-lactam resistance in nontypeable Haemophilus influenzae
- PMID: 24884375
- PMCID: PMC4039647
- DOI: 10.1186/1471-2180-14-131
Multilocus sequence typing and ftsI sequencing: a powerful tool for surveillance of penicillin-binding protein 3-mediated beta-lactam resistance in nontypeable Haemophilus influenzae
Abstract
Background: Beta-lactam resistance in Haemophilus influenzae due to ftsI mutations causing altered penicillin-binding protein 3 (PBP3) is increasing worldwide. Low-level resistant isolates with the N526K substitution (group II low-rPBP3) predominate in most geographical regions, while high-level resistant isolates with the additional S385T substitution (group III high-rPBP3) are common in Japan and South Korea.Knowledge about the molecular epidemiology of rPBP3 strains is limited. We combined multilocus sequence typing (MLST) and ftsI/PBP3 typing to study the emergence and spread of rPBP3 in nontypeable H. influenzae (NTHi) in Norway.
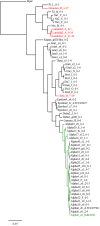
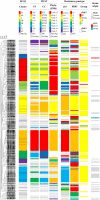
Results: The prevalence of rPBP3 in a population of 795 eye, ear and respiratory isolates (99% NTHi) from 2007 was 15%. The prevalence of clinical PBP3-mediated resistance to ampicillin was 9%, compared to 2.5% three years earlier. Group II low-rPBP3 predominated (96%), with significant proportions of isolates non-susceptible to cefotaxime (6%) and meropenem (20%). Group III high-rPBP3 was identified for the first time in Northern Europe.Four MLST sequence types (ST) with characteristic, highly diverging ftsI alleles accounted for 61% of the rPBP3 isolates. The most prevalent substitution pattern (PBP3 type A) was present in 41% of rPBP3 isolates, mainly carried by ST367 and ST14. Several unrelated STs possessed identical copies of the ftsI allele encoding PBP3 type A.Infection sites, age groups, hospitalization rates and rPBP3 frequencies differed between STs and phylogenetic groups.
Conclusions: This study is the first to link ftsI alleles to STs in H. influenzae. The results indicate that horizontal gene transfer contributes to the emergence of rPBP3 by phylogeny restricted transformation.Clonally related virulent rPBP3 strains are widely disseminated and high-level resistant isolates emerge in new geographical regions, threatening current empiric antibiotic treatment. The need of continuous monitoring of beta-lactam susceptibility and a global system for molecular surveillance of rPBP3 strains is underlined. Combining MLST and ftsI/PBP3 typing is a powerful tool for this purpose.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

