Construction and verification of the transcriptional regulatory response network of Streptococcus mutans upon treatment with the biofilm inhibitor carolacton
- PMID: 24884510
- PMCID: PMC4048456
- DOI: 10.1186/1471-2164-15-362
Construction and verification of the transcriptional regulatory response network of Streptococcus mutans upon treatment with the biofilm inhibitor carolacton
Erratum in
- BMC Genomics. 2014;15:739. Dobler, Irene W [corrected to Wagner-Döbler, Irene]
Abstract
Background: Carolacton is a newly identified secondary metabolite causing altered cell morphology and death of Streptococcus mutans biofilm cells. To unravel key regulators mediating these effects, the transcriptional regulatory response network of S. mutans biofilms upon carolacton treatment was constructed and analyzed. A systems biological approach integrating time-resolved transcriptomic data, reverse engineering, transcription factor binding sites, and experimental validation was carried out.
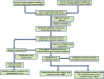
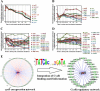
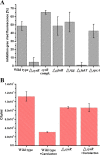
Results: The co-expression response network constructed from transcriptomic data using the reverse engineering algorithm called the Trend Correlation method consisted of 8284 gene pairs. The regulatory response network inferred by superimposing transcription factor binding site information into the co-expression network comprised 329 putative transcriptional regulatory interactions and could be classified into 27 sub-networks each co-regulated by a transcription factor. These sub-networks were significantly enriched with genes sharing common functions. The regulatory response network displayed global hierarchy and network motifs as observed in model organisms. The sub-networks modulated by the pyrimidine biosynthesis regulator PyrR, the glutamine synthetase repressor GlnR, the cysteine metabolism regulator CysR, global regulators CcpA and CodY and the two component system response regulators VicR and MbrC among others could putatively be related to the physiological effect of carolacton. The predicted interactions from the regulatory network between MbrC, known to be involved in cell envelope stress response, and the murMN-SMU_718c genes encoding peptidoglycan biosynthetic enzymes were experimentally confirmed using Electro Mobility Shift Assays. Furthermore, gene deletion mutants of five predicted key regulators from the response networks were constructed and their sensitivities towards carolacton were investigated. Deletion of cysR, the node having the highest connectivity among the regulators chosen from the regulatory network, resulted in a mutant which was insensitive to carolacton thus demonstrating not only the essentiality of cysR for the response of S. mutans biofilms to carolacton but also the relevance of the predicted network.
Conclusion: The network approach used in this study revealed important regulators and interactions as part of the response mechanisms of S. mutans biofilm cells to carolacton. It also opens a door for further studies into novel drug targets against streptococci.
Figures




Similar articles
-
The biofilm inhibitor carolacton disturbs membrane integrity and cell division of Streptococcus mutans through the serine/threonine protein kinase PknB.J Bacteriol. 2011 Oct;193(20):5692-706. doi: 10.1128/JB.05424-11. Epub 2011 Aug 12. J Bacteriol. 2011. PMID: 21840978 Free PMC article.
-
Damage of Streptococcus mutans biofilms by carolacton, a secondary metabolite from the myxobacterium Sorangium cellulosum.BMC Microbiol. 2010 Jul 26;10:199. doi: 10.1186/1471-2180-10-199. BMC Microbiol. 2010. PMID: 20659313 Free PMC article.
-
The biofilm inhibitor Carolacton inhibits planktonic growth of virulent pneumococci via a conserved target.Sci Rep. 2016 Jul 11;6:29677. doi: 10.1038/srep29677. Sci Rep. 2016. PMID: 27404808 Free PMC article.
-
Engineering of global transcription factors in Bacillus, a genetic tool for increasing product yields: a bioprocess overview.World J Microbiol Biotechnol. 2022 Nov 14;39(1):12. doi: 10.1007/s11274-022-03460-9. World J Microbiol Biotechnol. 2022. PMID: 36372802 Review.
-
General trends in the evolution of prokaryotic transcriptional regulatory networks.Genome Dyn. 2007;3:66-80. doi: 10.1159/000107604. Genome Dyn. 2007. PMID: 18753785 Review.
Cited by
-
Streptococcus mutans copes with heat stress by multiple transcriptional regulons modulating virulence and energy metabolism.Sci Rep. 2015 Aug 7;5:12929. doi: 10.1038/srep12929. Sci Rep. 2015. PMID: 26251057 Free PMC article.
-
Guarding the walls: the multifaceted roles of Bce modules in cell envelope stress sensing and antimicrobial resistance.J Bacteriol. 2024 Jul 25;206(7):e0012324. doi: 10.1128/jb.00123-24. Epub 2024 Jun 13. J Bacteriol. 2024. PMID: 38869304 Free PMC article. Review.
-
Combatting resistance: natural products as tools to drive the discovery of untapped antibiotic targets.Chem Commun (Camb). 2025 Aug 22. doi: 10.1039/d5cc03863d. Online ahead of print. Chem Commun (Camb). 2025. PMID: 40844298 Free PMC article. Review.
-
Carolacton Treatment Causes Delocalization of the Cell Division Proteins PknB and DivIVa in Streptococcus mutans in vivo.Front Microbiol. 2016 May 11;7:684. doi: 10.3389/fmicb.2016.00684. eCollection 2016. Front Microbiol. 2016. PMID: 27242711 Free PMC article.
-
Stress Physiology of Lactic Acid Bacteria.Microbiol Mol Biol Rev. 2016 Jul 27;80(3):837-90. doi: 10.1128/MMBR.00076-15. Print 2016 Sep. Microbiol Mol Biol Rev. 2016. PMID: 27466284 Free PMC article. Review.
References
-
- Dufour D, Leung V, Levesque CM. Bacterial biofilm: structure, function, and antimicrobial resistance. Endo Topics. 2012;22(1):2–16. doi: 10.1111/j.1601-1546.2012.00277.x. - DOI
-
- Roychoudhury S, Zielinski NA, Ninfa AJ, Allen NE, Jungheim LN, Nicas TI, Chakrabarty AM. Inhibitors of two-component signal transduction systems: inhibition of alginate gene activation in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1993;90:965–969. doi: 10.1073/pnas.90.3.965. - DOI - PMC - PubMed
-
- Reck M, Rutz K, Kunze B, Tomasch J, Surapaneni SK, Schulz S, Wagner-Döbler I. The biofilm inhibitor carolacton disturbs membrane integrity and cell division of Streptococcus mutans through the serine/threonine protein kinase PknB. J Bacteriol. 2011;193:5692–5706. doi: 10.1128/JB.05424-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

