Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids
- PMID: 24884625
- PMCID: PMC4045975
- DOI: 10.1186/1471-2164-15-405
Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids
Abstract
Background: Rhazya stricta is native to arid regions in South Asia and the Middle East and is used extensively in folk medicine to treat a wide range of diseases. In addition to generating genomic resources for this medicinally important plant, analyses of the complete plastid and mitochondrial genomes and a nuclear transcriptome from Rhazya provide insights into inter-compartmental transfers between genomes and the patterns of evolution among eight asterid mitochondrial genomes.
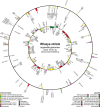
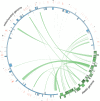
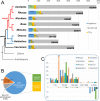
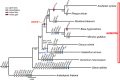
Results: The 154,841 bp plastid genome is highly conserved with gene content and order identical to the ancestral organization of angiosperms. The 548,608 bp mitochondrial genome exhibits a number of phenomena including the presence of recombinogenic repeats that generate a multipartite organization, transferred DNA from the plastid and nuclear genomes, and bidirectional DNA transfers between the mitochondrion and the nucleus. The mitochondrial genes sdh3 and rps14 have been transferred to the nucleus and have acquired targeting presequences. In the case of rps14, two copies are present in the nucleus; only one has a mitochondrial targeting presequence and may be functional. Phylogenetic analyses of both nuclear and mitochondrial copies of rps14 across angiosperms suggests Rhazya has experienced a single transfer of this gene to the nucleus, followed by a duplication event. Furthermore, the phylogenetic distribution of gene losses and the high level of sequence divergence in targeting presequences suggest multiple, independent transfers of both sdh3 and rps14 across asterids. Comparative analyses of mitochondrial genomes of eight sequenced asterids indicates a complicated evolutionary history in this large angiosperm clade with considerable diversity in genome organization and size, repeat, gene and intron content, and amount of foreign DNA from the plastid and nuclear genomes.
Conclusions: Organelle genomes of Rhazya stricta provide valuable information for improving the understanding of mitochondrial genome evolution among angiosperms. The genomic data have enabled a rigorous examination of the gene transfer events. Rhazya is unique among the eight sequenced asterids in the types of events that have shaped the evolution of its mitochondrial genome. Furthermore, the organelle genomes of R. stricta provide valuable genomic resources for utilizing this important medicinal plant in biotechnology applications.
Figures




Similar articles
-
Insights into the nuclear-organelle DNA integration in Cicuta virosa (Apiaceae) provided by complete plastid and mitochondrial genomes.BMC Genomics. 2025 Feb 3;26(1):102. doi: 10.1186/s12864-025-11230-8. BMC Genomics. 2025. PMID: 39901091 Free PMC article.
-
Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae.BMC Plant Biol. 2015 Feb 5;15:40. doi: 10.1186/s12870-015-0432-6. BMC Plant Biol. 2015. PMID: 25652741 Free PMC article.
-
Complete plastid genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms.BMC Genomics. 2006 Aug 31;7:222. doi: 10.1186/1471-2164-7-222. BMC Genomics. 2006. PMID: 16945140 Free PMC article.
-
Biotechnology of the medicinal plant Rhazya stricta: a little investigated member of the Apocynaceae family.Biotechnol Lett. 2017 Jun;39(6):829-840. doi: 10.1007/s10529-017-2320-7. Epub 2017 Mar 15. Biotechnol Lett. 2017. PMID: 28299544 Review.
-
Piece and parcel of gymnosperm organellar genomes.Planta. 2024 Jun 3;260(1):14. doi: 10.1007/s00425-024-04449-4. Planta. 2024. PMID: 38829418 Review.
Cited by
-
Mitochondrial plastid DNA can cause DNA barcoding paradox in plants.Sci Rep. 2020 Apr 9;10(1):6112. doi: 10.1038/s41598-020-63233-y. Sci Rep. 2020. PMID: 32273595 Free PMC article.
-
Genomics and Evolution in Traditional Medicinal Plants: Road to a Healthier Life.Evol Bioinform Online. 2015 Oct 4;11:197-212. doi: 10.4137/EBO.S31326. eCollection 2015. Evol Bioinform Online. 2015. PMID: 26461812 Free PMC article. Review.
-
Characterization of Chloroplast Genomes From Two Salvia Medicinal Plants and Gene Transfer Among Their Mitochondrial and Chloroplast Genomes.Front Genet. 2020 Oct 22;11:574962. doi: 10.3389/fgene.2020.574962. eCollection 2020. Front Genet. 2020. PMID: 33193683 Free PMC article.
-
Intrageneric structural variation in organelle genomes from the genus Dystaenia (Apiaceae): genome rearrangement and mitochondrion-to-plastid DNA transfer.Front Plant Sci. 2023 Dec 5;14:1283292. doi: 10.3389/fpls.2023.1283292. eCollection 2023. Front Plant Sci. 2023. PMID: 38116150 Free PMC article.
-
Comparative analysis of mitochondrial genomes of two alpine medicinal plants of Gentiana (Gentianaceae).PLoS One. 2023 Jan 26;18(1):e0281134. doi: 10.1371/journal.pone.0281134. eCollection 2023. PLoS One. 2023. PMID: 36701356 Free PMC article.
References
-
- Ageel AM, Mossa JS, Al-Yahya MA, Tariq M, Al-Said MS. Plants Used in Saudi Folk Medicine. Riyadh, Saudia Arabia: King Saud University Press; 1987.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- SRA/SRR1151604
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

