Automated ensemble assembly and validation of microbial genomes
- PMID: 24884846
- PMCID: PMC4030574
- DOI: 10.1186/1471-2105-15-126
Automated ensemble assembly and validation of microbial genomes
Abstract
Background: The continued democratization of DNA sequencing has sparked a new wave of development of genome assembly and assembly validation methods. As individual research labs, rather than centralized centers, begin to sequence the majority of new genomes, it is important to establish best practices for genome assembly. However, recent evaluations such as GAGE and the Assemblathon have concluded that there is no single best approach to genome assembly. Instead, it is preferable to generate multiple assemblies and validate them to determine which is most useful for the desired analysis; this is a labor-intensive process that is often impossible or unfeasible.
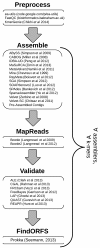
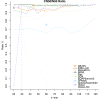
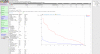
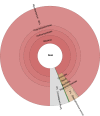
Results: To encourage best practices supported by the community, we present iMetAMOS, an automated ensemble assembly pipeline; iMetAMOS encapsulates the process of running, validating, and selecting a single assembly from multiple assemblies. iMetAMOS packages several leading open-source tools into a single binary that automates parameter selection and execution of multiple assemblers, scores the resulting assemblies based on multiple validation metrics, and annotates the assemblies for genes and contaminants. We demonstrate the utility of the ensemble process on 225 previously unassembled Mycobacterium tuberculosis genomes as well as a Rhodobacter sphaeroides benchmark dataset. On these real data, iMetAMOS reliably produces validated assemblies and identifies potential contamination without user intervention. In addition, intelligent parameter selection produces assemblies of R. sphaeroides comparable to or exceeding the quality of those from the GAGE-B evaluation, affecting the relative ranking of some assemblers.
Conclusions: Ensemble assembly with iMetAMOS provides users with multiple, validated assemblies for each genome. Although computationally limited to small or mid-sized genomes, this approach is the most effective and reproducible means for generating high-quality assemblies and enables users to select an assembly best tailored to their specific needs.
Figures

References
-
- Bradnam K, Fass J, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert S, Chapman J, Chapuis G, Chikhi R, Chitsaz H, Chou W-C, Corbeil J, Del Fabbro C, Docking T, Durbin R, Earl D, Emrich S, Fedotov P, Fonseca N, Ganapathy G, Gibbs R, Gnerre S, Godzaridis E, Goldstein S, Haimel M, Hall G, Haussler D, Hiatt J, Ho I. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience. 2013;2(1):10. doi: 10.1186/2047-217X-2-10. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

