The chemical arsenal of Burkholderia pseudomallei is essential for pathogenicity
- PMID: 24884988
- PMCID: PMC4091270
- DOI: 10.1021/ja504617n
The chemical arsenal of Burkholderia pseudomallei is essential for pathogenicity
Abstract
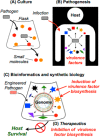
Increasing evidence has shown that small-molecule chemistry in microbes (i.e., secondary metabolism) can modulate the microbe-host response in infection and pathogenicity. The bacterial disease melioidosis is conferred by the highly virulent, antibiotic-resistant pathogen Burkholderia pseudomallei (BP). Whereas some macromolecular structures have been shown to influence BP virulence (e.g., secretion systems, cellular capsule, pili), the role of the large cryptic secondary metabolome encoded within its genome has been largely unexplored for its importance to virulence. Herein we demonstrate that BP-encoded small-molecule biosynthesis is indispensible for in vivo BP pathogenicity. Promoter exchange experiments were used to induce high-level molecule production from two gene clusters (MPN and SYR) found to be essential for in vivo virulence. NMR structural characterization of these metabolites identified a new class of lipopeptide biosurfactants/biofilm modulators (the malleipeptins) and syrbactin-type proteasome inhibitors, both of which represent overlooked small-molecule virulence factors for BP. Disruption of Burkholderia virulence by inhibiting the biosynthesis of these small-molecule biosynthetic pathways may prove to be an effective strategy for developing novel melioidosis-specific therapeutics.
Figures
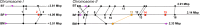




References
-
- Galyov E. E.; Brett P. J.; DeShazer D. Annu. Rev. Microbiol. 2010, 64, 495. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

