Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing
- PMID: 24885028
- PMCID: PMC4032349
- DOI: 10.1186/1755-8794-7-23
Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing
Abstract
Background: Clinical specimens undergoing diagnostic molecular pathology testing are fixed in formalin due to the necessity for detailed morphological assessment. However, formalin fixation can cause major issues with molecular testing, as it causes DNA damage such as fragmentation and non-reproducible sequencing artefacts after PCR amplification. In the context of massively parallel sequencing (MPS), distinguishing true low frequency variants from sequencing artefacts remains challenging. The prevalence of formalin-induced DNA damage and its impact on molecular testing and clinical genomics remains poorly understood.
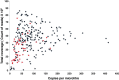
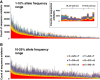
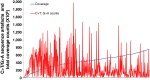
Methods: The Cancer 2015 study is a population-based cancer cohort used to assess the feasibility of mutational screening using MPS in cancer patients from Victoria, Australia. While blocks were formalin-fixed and paraffin-embedded in different anatomical pathology laboratories, they were centrally extracted for DNA utilising the same protocol, and run through the same MPS platform (Illumina TruSeq Amplicon Cancer Panel). The sequencing artefacts in the 1-10% and the 10-25% allele frequency ranges were assessed in 488 formalin-fixed tumours from the pilot phase of the Cancer 2015 cohort. All blocks were less than 2.5 years of age (mean 93 days).
Results: Consistent with the signature of DNA damage due to formalin fixation, many formalin-fixed samples displayed disproportionate levels of C>T/G>A changes in the 1-10% allele frequency range. Artefacts were less apparent in the 10-25% allele frequency range. Significantly, changes were inversely correlated with coverage indicating high levels of sequencing artefacts were associated with samples with low amounts of available amplifiable template due to fragmentation. The degree of fragmentation and sequencing artefacts differed between blocks sourced from different anatomical pathology laboratories. In a limited validation of potentially actionable low frequency mutations, a NRAS G12D mutation in a melanoma was shown to be a false positive.
Conclusions: These findings indicate that DNA damage following formalin fixation remains a major challenge in laboratories working with MPS. Methodologies that assess, minimise or remove formalin-induced DNA damaged templates as part of MPS protocols will aid in the interpretation of genomic results leading to better patient outcomes.
Figures

Similar articles
-
Reducing sequence artifacts in amplicon-based massively parallel sequencing of formalin-fixed paraffin-embedded DNA by enzymatic depletion of uracil-containing templates.Clin Chem. 2013 Sep;59(9):1376-83. doi: 10.1373/clinchem.2012.202390. Epub 2013 May 6. Clin Chem. 2013. PMID: 23649127
-
The efficacy of uracil DNA glycosylase pretreatment in amplicon-based massively parallel sequencing with DNA extracted from archived formalin-fixed paraffin-embedded esophageal cancer tissues.Cancer Genet. 2015 Sep;208(9):415-27. doi: 10.1016/j.cancergen.2015.05.001. Epub 2015 May 11. Cancer Genet. 2015. PMID: 26194062
-
Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil- DNA glycosylase.Oncotarget. 2012 May;3(5):546-58. doi: 10.18632/oncotarget.503. Oncotarget. 2012. PMID: 22643842 Free PMC article.
-
Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization.Clin Chem. 2015 Jan;61(1):64-71. doi: 10.1373/clinchem.2014.223040. Epub 2014 Nov 24. Clin Chem. 2015. PMID: 25421801 Review.
-
Genome-scale mutational signature analysis in archived fixed tissues.Mutat Res Rev Mutat Res. 2024 Jul-Dec;794:108512. doi: 10.1016/j.mrrev.2024.108512. Epub 2024 Aug 29. Mutat Res Rev Mutat Res. 2024. PMID: 39216514 Review.
Cited by
-
Mutational analysis of primary and metastatic colorectal cancer samples underlying the resistance to cetuximab-based therapy.Onco Targets Ther. 2016 Jul 28;9:4695-703. doi: 10.2147/OTT.S102891. eCollection 2016. Onco Targets Ther. 2016. PMID: 27555788 Free PMC article.
-
Integration of Wet and Dry Bench Processes Optimizes Targeted Next-generation Sequencing of Low-quality and Low-quantity Tumor Biopsies.J Vis Exp. 2016 Apr 11;(110):e53836. doi: 10.3791/53836. J Vis Exp. 2016. PMID: 27166994 Free PMC article.
-
A novel approach to stabilize fetal cell-free DNA fraction in maternal blood samples for extended period of time.PLoS One. 2018 Dec 6;13(12):e0208508. doi: 10.1371/journal.pone.0208508. eCollection 2018. PLoS One. 2018. PMID: 30521613 Free PMC article.
-
Understanding the Mechanisms of Resistance in EGFR-Positive NSCLC: From Tissue to Liquid Biopsy to Guide Treatment Strategy.Int J Mol Sci. 2019 Aug 14;20(16):3951. doi: 10.3390/ijms20163951. Int J Mol Sci. 2019. PMID: 31416192 Free PMC article. Review.
-
Systematic evaluation of PAXgene® tissue fixation for the histopathological and molecular study of lung cancer.J Pathol Clin Res. 2020 Jan;6(1):40-54. doi: 10.1002/cjp2.145. Epub 2019 Nov 11. J Pathol Clin Res. 2020. PMID: 31571426 Free PMC article.
References
-
- MacConaill LE, Campbell CD, Kehoe SM, Bass AJ, Hatton C, Niu L, Davis M, Yao K, Hanna M, Mondal C, Luongo L, Emery CM, Baker AC, Philips J, Goff DJ, Fiorentino M, Rubin MA, Polyak K, Chan J, Wang Y, Fletcher JA, Santagata S, Corso G, Roviello F, Shivdasani R, Kieran MW, Ligon KL, Stiles CD, Hahn WC, Meyerson ML. et al.Profiling critical cancer gene mutations in clinical tumor samples. PloS one. 2009;7:e7887. doi: 10.1371/journal.pone.0007887. - DOI - PMC - PubMed
-
- Mar VJ, Wong SQ, Li J, Scolyer RA, McLean C, Papenfuss AT, Tothill RW, Kakavand H, Mann GJ, Thompson JF, Behren A, Cebon JS, Wolfe R, Kelly JW, Dobrovic A, McArthur GA. BRAF/NRAS wild-type melanomas have a high mutation load correlating with histologic and molecular signatures of UV damage. Clin Cancer Res. 2013;7:4589–4598. doi: 10.1158/1078-0432.CCR-13-0398. - DOI - PubMed
-
- Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, Ducar M, Van Hummelen P, Macconaill LE, Hahn WC, Meyerson M, Gabriel SB, Garraway LA. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer discovery. 2012;7(1):82–93. doi: 10.1158/2159-8290.CD-11-0184. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

