Complete steady-state rate equation for DNA ligase and its use for measuring product kinetic parameters of NAD⁺-dependent DNA ligase from Haemophilus influenzae
- PMID: 24885075
- PMCID: PMC4022432
- DOI: 10.1186/1756-0500-7-287
Complete steady-state rate equation for DNA ligase and its use for measuring product kinetic parameters of NAD⁺-dependent DNA ligase from Haemophilus influenzae
Abstract
Background: DNA ligase seals the nicks in the phosphodiester backbone between Okazaki fragments during DNA replication. DNA ligase has an unusual Bi Ter Ping Pong kinetic mechanism. Its substrates in eubacteria are NAD+ and nicked DNA (nDNA). Its products are nicotinamide mononucleotide (NMN), adenosine 5'-monophosphate (AMP), and sealed DNA. Investigation of the kinetic mechanism and measurement of the kinetic constants of DNA ligase using steady-state kinetics would benefit from the availability of the complete steady-state rate equation, including terms for product concentrations and product-related kinetic constants, which has not previously been published.
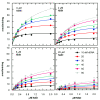
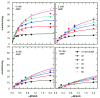
Results: The rate equations for two possible Bi Ter kinetic mechanisms for DNA ligase, including products, are reported. The mechanisms differ according to whether the last two products, AMP and sealed DNA, are released in an ordered or rapid-equilibrium random (RER) manner. Steady-state kinetic studies of product inhibition by NMN and AMP were performed with Haemophilus influenzae NAD+-dependent DNA ligase. The complete rate equation enabled measurement of dissociation constants for NAD+, NMN, and AMP and eliminated one of 3 possible product release mechanisms.
Conclusions: Steady-state kinetic product inhibition experiments and complete steady-state kinetic rate equations were used to measure dissociation constants of NAD+, NMN, and AMP and eliminate the possibility that AMP is the second product released in an ordered mechanism. Determining by steady-state kinetics whether the release of sealed DNA and AMP products goes by an ordered (AMP last off) or RER mechanism was shown to require a product inhibition study using sealed DNA.
Figures

Similar articles
-
A high-throughput fluorescence resonance energy transfer-based assay for DNA ligase.J Biomol Screen. 2011 Jun;16(5):486-93. doi: 10.1177/1087057111398295. Epub 2011 Mar 11. J Biomol Screen. 2011. PMID: 21398623
-
NadN and e (P4) are essential for utilization of NAD and nicotinamide mononucleotide but not nicotinamide riboside in Haemophilus influenzae.J Bacteriol. 2001 Jul;183(13):3974-81. doi: 10.1128/JB.183.13.3974-3981.2001. J Bacteriol. 2001. PMID: 11395461 Free PMC article.
-
Rapid Time Scale Analysis of T4 DNA Ligase-DNA Binding.Biochemistry. 2017 Feb 28;56(8):1117-1129. doi: 10.1021/acs.biochem.6b01261. Epub 2017 Feb 16. Biochemistry. 2017. PMID: 28165732
-
DNA and RNA ligases: structural variations and shared mechanisms.Curr Opin Struct Biol. 2008 Feb;18(1):96-105. doi: 10.1016/j.sbi.2007.12.008. Epub 2008 Feb 8. Curr Opin Struct Biol. 2008. PMID: 18262407 Review.
-
DNA ligase: structure, mechanism, and function.Science. 1974 Nov 29;186(4166):790-7. doi: 10.1126/science.186.4166.790. Science. 1974. PMID: 4377758 Review.
Cited by
-
Biochemical and Structural Characterisation of DNA Ligases from Bacteria and Archaea.Biosci Rep. 2016 Oct 6;36(5):00391. doi: 10.1042/BSR20160003. Biosci Rep. 2016. PMID: 27582505 Free PMC article.
-
The Inhibitory Effect of Non-Substrate and Substrate DNA on the Ligation and Self-Adenylylation Reactions Catalyzed by T4 DNA Ligase.PLoS One. 2016 Mar 8;11(3):e0150802. doi: 10.1371/journal.pone.0150802. eCollection 2016. PLoS One. 2016. PMID: 26954034 Free PMC article.
-
Kinetic mechanism and fidelity of nick sealing by Escherichia coli NAD+-dependent DNA ligase (LigA).Nucleic Acids Res. 2016 Mar 18;44(5):2298-309. doi: 10.1093/nar/gkw049. Epub 2016 Feb 8. Nucleic Acids Res. 2016. PMID: 26857547 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

