In silico analyses reveal common cellular pathways affected by loss of heterozygosity (LOH) events in the lymphomagenesis of Non-Hodgkin's lymphoma (NHL)
- PMID: 24885312
- PMCID: PMC4041994
- DOI: 10.1186/1471-2164-15-390
In silico analyses reveal common cellular pathways affected by loss of heterozygosity (LOH) events in the lymphomagenesis of Non-Hodgkin's lymphoma (NHL)
Abstract
Background: The analysis of cellular networks and pathways involved in oncogenesis has increased our knowledge about the pathogenic mechanisms that underlie tumour biology and has unmasked new molecular targets that may lead to the design of better anti-cancer therapies. Recently, using a high resolution loss of heterozygosity (LOH) analysis, we identified a number of potential tumour suppressor genes (TSGs) within common LOH regions across cases suffering from two of the most common forms of Non-Hodgkin's lymphoma (NHL), Follicular Lymphoma (FL) and Diffuse Large B-cell Lymphoma (DLBCL). From these studies LOH of the protein tyrosine phosphatase receptor type J (PTPRJ) gene was identified as a common event in the lymphomagenesis of these B-cell lymphomas. The present study aimed to determine the cellular pathways affected by the inactivation of these TSGs including PTPRJ in FL and DLBCL tumourigenesis.
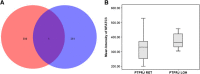
Results: Pathway analytical approaches identified that candidate TSGs located within common LOH regions participate within cellular pathways, which may play a crucial role in FL and DLBCL lymphomagenesis (i.e., metabolic pathways). These analyses also identified genes within the interactome of PTPRJ (i.e. PTPN11 and B2M) that when inactivated in NHL may play an important role in tumourigenesis. We also detected genes that are differentially expressed in cases with and without LOH of PTPRJ, such as NFATC3 (nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3). Moreover, upregulation of the VEGF, MAPK and ERBB signalling pathways was also observed in NHL cases with LOH of PTPRJ, indicating that LOH-driving events causing inactivation of PTPRJ, apart from possibly inducing a constitutive activation of these pathways by reduction or abrogation of its dephosphorylation activity, may also induce upregulation of these pathways when inactivated. This finding implicates these pathways in the lymphomagenesis and progression of FL and DLBCL.
Conclusions: The evidence obtained in this research supports findings suggesting that FL and DLBCL share common pathogenic mechanisms. Also, it indicates that PTPRJ can play a crucial role in the pathogenesis of these B-cell tumours and suggests that activation of PTPRJ might be an interesting novel chemotherapeutic target for the treatment of these B-cell tumours.
Figures



Similar articles
-
High-resolution loss of heterozygosity screening implicates PTPRJ as a potential tumor suppressor gene that affects susceptibility to Non-Hodgkin's lymphoma.Genes Chromosomes Cancer. 2013 May;52(5):467-79. doi: 10.1002/gcc.22044. Epub 2013 Jan 23. Genes Chromosomes Cancer. 2013. PMID: 23341091
-
A new method to detect loss of heterozygosity using cohort heterozygosity comparisons.BMC Cancer. 2010 May 12;10:195. doi: 10.1186/1471-2407-10-195. BMC Cancer. 2010. PMID: 20462409 Free PMC article.
-
The p73 locus is commonly deleted in non-Hodgkin's lymphomas.Leuk Res. 2004 Dec;28(12):1341-5. doi: 10.1016/j.leukres.2004.04.010. Leuk Res. 2004. PMID: 15475076
-
Pathogenesis of non-Hodgkin's lymphoma.J Clin Oncol. 2011 May 10;29(14):1803-11. doi: 10.1200/JCO.2010.33.3252. Epub 2011 Apr 11. J Clin Oncol. 2011. PMID: 21483013 Review.
-
Non-Hodgkin's lymphoma: the old and the new.Clin Lymphoma Myeloma Leuk. 2011 Jun;11 Suppl 1:S87-90. doi: 10.1016/j.clml.2011.03.029. Epub 2011 Apr 28. Clin Lymphoma Myeloma Leuk. 2011. PMID: 22035756 Review.
Cited by
-
ATP-binding cassette member B5 (ABCB5) promotes tumor cell invasiveness in human colorectal cancer.J Biol Chem. 2018 Jul 13;293(28):11166-11178. doi: 10.1074/jbc.RA118.003187. Epub 2018 May 22. J Biol Chem. 2018. PMID: 29789423 Free PMC article.
-
Loss of function mutations in PTPN6 promote STAT3 deregulation via JAK3 kinase in diffuse large B-cell lymphoma.Oncotarget. 2015 Dec 29;6(42):44703-13. doi: 10.18632/oncotarget.6300. Oncotarget. 2015. PMID: 26565811 Free PMC article.
-
Whole genome sequencing analysis of high confidence variants of B-cell lymphoma in Canis familiaris.PLoS One. 2020 Aug 28;15(8):e0238183. doi: 10.1371/journal.pone.0238183. eCollection 2020. PLoS One. 2020. PMID: 32857815 Free PMC article.
-
The Structure, Function and Regulation of Protein Tyrosine Phosphatase Receptor Type J and Its Role in Diseases.Cells. 2022 Dec 20;12(1):8. doi: 10.3390/cells12010008. Cells. 2022. PMID: 36611803 Free PMC article. Review.
-
Alpha5 Nicotinic Acetylcholine Receptor Contributes to Nicotine-Induced Lung Cancer Development and Progression.Front Pharmacol. 2017 Aug 23;8:573. doi: 10.3389/fphar.2017.00573. eCollection 2017. Front Pharmacol. 2017. PMID: 28878681 Free PMC article.
References
-
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer (IARC); 2008.
-
- Turner JJ, Morton LM, Linet MS, Clarke CA, Kadin ME, Vajdic CM, Monnereau A, Maynadie M, Chiu BC, Marcos-Gragera R, Costantini AS, Cerhan JR, Weisenburger DD. InterLymph hierarchical classification of lymphoid neoplasms for epidemiologic research based on the WHO classification (2008): update and future directions. Blood. 2010;116(20):e90–98. doi: 10.1182/blood-2010-06-289561. - DOI - PMC - PubMed
-
- Jayasekara H, Karahalios A, Juneja S, Thursfield V, Farrugia H, English DR, Giles GG. Incidence and survival of lymphohematopoietic neoplasms according to the World Health Organization classification: a population-based study from the Victorian Cancer Registry in Australia. Leuk Lymphoma. 2010;51(3):456–468. doi: 10.3109/10428190903552104. - DOI - PubMed
-
- Federico M, Luminari S, Dondi A, Tucci A, Vitolo U, Rigacci L, Di Raimondo F, Carella AM, Pulsoni A, Merli F, Arcaini L, Angrilli F, Stelitano C, Gaidano G, Dell'Olio M, Marcheselli L, Franco V, Galimberti S, Sacchi S, Brugiatelli M. R-CVP Versus R-CHOP Versus R-FM for the initial treatment of patients with advanced-stage follicular lymphoma: results of the FOLL05 trial conducted by the Fondazione Italiana Linfomi. J Clin Oncol. 2013;31:1506–1513. doi: 10.1200/JCO.2012.45.0866. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

