Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor
- PMID: 24885329
- PMCID: PMC4068068
- DOI: 10.1186/1741-7007-12-41
Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor
Abstract
Background: Klebsiella pneumoniae strains are pathogenic to animals and humans, in which they are both a frequent cause of nosocomial infections and a re-emerging cause of severe community-acquired infections. K. pneumoniae isolates of the capsular serotype K2 are among the most virulent. In order to identify novel putative virulence factors that may account for the severity of K2 infections, the genome sequence of the K2 reference strain Kp52.145 was determined and compared to two K1 and K2 strains of low virulence and to the reference strains MGH 78578 and NTUH-K2044.
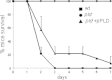
Results: In addition to diverse functions related to host colonization and virulence encoded in genomic regions common to the four strains, four genomic islands specific for Kp52.145 were identified. These regions encoded genes for the synthesis of colibactin toxin, a putative cytotoxin outer membrane protein, secretion systems, nucleases and eukaryotic-like proteins. In addition, an insertion within a type VI secretion system locus included sel1 domain containing proteins and a phospholipase D family protein (PLD1). The pld1 mutant was avirulent in a pneumonia model in mouse. The pld1 mRNA was expressed in vivo and the pld1 gene was associated with K. pneumoniae isolates from severe infections. Analysis of lipid composition of a defective E. coli strain complemented with pld1 suggests an involvement of PLD1 in cardiolipin metabolism.
Conclusions: Determination of the complete genome of the K2 reference strain identified several genomic islands comprising putative elements of pathogenicity. The role of PLD1 in pathogenesis was demonstrated for the first time and suggests that lipid metabolism is a novel virulence mechanism of K. pneumoniae.
Figures




Similar articles
-
Correlation of Klebsiella pneumoniae comparative genetic analyses with virulence profiles in a murine respiratory disease model.PLoS One. 2014 Sep 9;9(9):e107394. doi: 10.1371/journal.pone.0107394. eCollection 2014. PLoS One. 2014. PMID: 25203254 Free PMC article.
-
Whole-Genome-Sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374.Virulence. 2018 Jan 1;9(1):510-521. doi: 10.1080/21505594.2017.1421894. Virulence. 2018. PMID: 29338592 Free PMC article.
-
Mapping the Evolution of Hypervirulent Klebsiella pneumoniae.mBio. 2015 Jul 21;6(4):e00630. doi: 10.1128/mBio.00630-15. mBio. 2015. PMID: 26199326 Free PMC article.
-
Clinical Implications of Genomic Adaptation and Evolution of Carbapenem-Resistant Klebsiella pneumoniae.J Infect Dis. 2017 Feb 15;215(suppl_1):S18-S27. doi: 10.1093/infdis/jiw378. J Infect Dis. 2017. PMID: 28375514 Free PMC article. Review.
-
Modified Impedance Sensing System Determination of Virulence Characteristics of Pathogenic Bacteria Klebsiella Species.Indian J Microbiol. 2023 Dec;63(4):421-428. doi: 10.1007/s12088-023-01112-6. Epub 2023 Oct 27. Indian J Microbiol. 2023. PMID: 38031597 Free PMC article. Review.
Cited by
-
Current status and development prospects of aquatic vaccines.Front Immunol. 2022 Nov 10;13:1040336. doi: 10.3389/fimmu.2022.1040336. eCollection 2022. Front Immunol. 2022. PMID: 36439092 Free PMC article. Review.
-
Virulence genes and previously unexplored gene clusters in four commensal Neisseria spp. isolated from the human throat expand the neisserial gene repertoire.Microb Genom. 2020 Sep;6(9):mgen000423. doi: 10.1099/mgen.0.000423. Epub 2020 Aug 26. Microb Genom. 2020. PMID: 32845827 Free PMC article.
-
Distribution and diversity of type VI secretion system clusters in Enterobacter bugandensis and Enterobacter cloacae.Microb Genom. 2023 Dec;9(12):001148. doi: 10.1099/mgen.0.001148. Microb Genom. 2023. PMID: 38054968 Free PMC article.
-
Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage.BMC Microbiol. 2018 Aug 29;18(1):94. doi: 10.1186/s12866-018-1236-2. BMC Microbiol. 2018. PMID: 30157774 Free PMC article.
-
Dissociation between the critical role of ClpB of Francisella tularensis for the heat shock response and the DnaK interaction and its important role for efficient type VI secretion and bacterial virulence.PLoS Pathog. 2020 Apr 10;16(4):e1008466. doi: 10.1371/journal.ppat.1008466. eCollection 2020 Apr. PLoS Pathog. 2020. PMID: 32275693 Free PMC article.
References
-
- Brisse S, Grimont F, Grimont PAD. The Prokaryotes: A Handbook on the Biology of Bacteria. 3. Vol. 12. New York: Springer NY; 2006. The genus Klebsiella; pp. 159–196.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

