Identification of candidate risk gene variations by whole-genome sequence analysis of four rat strains commonly used in inflammation research
- PMID: 24885425
- PMCID: PMC4041999
- DOI: 10.1186/1471-2164-15-391
Identification of candidate risk gene variations by whole-genome sequence analysis of four rat strains commonly used in inflammation research
Abstract
Background: The DA rat strain is particularly susceptible to the induction of a number of chronic inflammatory diseases, such as models for rheumatoid arthritis and multiple sclerosis. Here we sequenced the genomes of two DA sub-strains and two disease resistant strains, E3 and PVG, previously used together with DA strains in genetically segregating crosses.
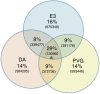
Results: The data uncovers genomic variations, such as single nucleotide variations (SNVs) and copy number variations that underlie phenotypic differences between the strains. Comparisons of regional differences between the two DA sub-strains identified 8 genomic regions that discriminate between the strains that together cover 38 Mbp and harbor 302 genes. We analyzed 10 fine-mapped quantitative trait loci and our data implicate strong candidates for genetic variations that mediate their effects. For example we could identify a single SNV candidate in a regulatory region of the gene Il21r, which has been associated to differential expression in both rats and human MS patients. In the APLEC complex we identified two SNVs in a highly conserved region, which could affect the regulation of all APLEC encoded genes and explain the polygenic differential expression seen in the complex. Furthermore, the non-synonymous SNV modifying aa153 of the Ncf1 protein was confirmed as the sole causative factor.
Conclusion: This complete map of genetic differences between the most commonly used rat strains in inflammation research constitutes an important reference in understanding how genetic variations contribute to the traits of importance for inflammatory diseases.
Figures


Similar articles
-
Definition of arthritis candidate risk genes by combining rat linkage-mapping results with human case-control association data.Ann Rheum Dis. 2009 Dec;68(12):1925-32. doi: 10.1136/ard.2008.090803. Epub 2008 Dec 9. Ann Rheum Dis. 2009. PMID: 19066175
-
Association of arthritis with a gene complex encoding C-type lectin-like receptors.Arthritis Rheum. 2007 Aug;56(8):2620-32. doi: 10.1002/art.22813. Arthritis Rheum. 2007. PMID: 17665455
-
Detection of arthritis-susceptibility loci, including Ncf1, and variable effects of the major histocompatibility complex region depending on genetic background in rats.Arthritis Rheum. 2009 Feb;60(2):419-27. doi: 10.1002/art.24292. Arthritis Rheum. 2009. PMID: 19180494
-
Dissection of the genetic complexity of arthritis using animal models.J Autoimmun. 2003 Sep;21(2):99-103. doi: 10.1016/s0896-8411(03)00096-9. J Autoimmun. 2003. PMID: 12935777 Review.
-
Quest for arthritis-causative genetic factors in the rat.Physiol Genomics. 2006 Oct 3;27(1):1-11. doi: 10.1152/physiolgenomics.00034.2005. Epub 2006 Jun 27. Physiol Genomics. 2006. PMID: 16804090 Review.
Cited by
-
Applications of Next-generation Sequencing in Systemic Autoimmune Diseases.Genomics Proteomics Bioinformatics. 2015 Aug;13(4):242-9. doi: 10.1016/j.gpb.2015.09.004. Epub 2015 Oct 15. Genomics Proteomics Bioinformatics. 2015. PMID: 26432094 Free PMC article. Review.
-
Rat models of human diseases and related phenotypes: a systematic inventory of the causative genes.J Biomed Sci. 2020 Aug 2;27(1):84. doi: 10.1186/s12929-020-00673-8. J Biomed Sci. 2020. PMID: 32741357 Free PMC article. Review.
-
Endophilin A2 deficiency protects rodents from autoimmune arthritis by modulating T cell activation.Nat Commun. 2021 Jan 27;12(1):610. doi: 10.1038/s41467-020-20586-2. Nat Commun. 2021. PMID: 33504785 Free PMC article.
-
Identification of Clec4b as a novel regulator of bystander activation of auto-reactive T cells and autoimmune disease.PLoS Genet. 2020 Jun 4;16(6):e1008788. doi: 10.1371/journal.pgen.1008788. eCollection 2020 Jun. PLoS Genet. 2020. PMID: 32497089 Free PMC article.
-
Genome-to-phenome research in rats: progress and perspectives.Int J Biol Sci. 2021 Jan 1;17(1):119-133. doi: 10.7150/ijbs.51628. eCollection 2021. Int J Biol Sci. 2021. PMID: 33390838 Free PMC article. Review.
References
-
- Atanur SS, Birol I, Guryev V, Hirst M, Hummel O, Morrissey C, Behmoaras J, Fernandez-Suarez XM, Johnson MD, McLaren WM, Patone G, Petretto E, Plessy C, Rockland KS, Rockland C, Saar K, Zhao Y, Carninci P, Flicek P, Kurtz T, Cuppen E, Pravenec M, Hubner N, Jones SJM, Birney E, Aitman TJ. The genome sequence of the spontaneously hypertensive rat: analysis and functional significance. Genome Res. 2010;20:791–803. doi: 10.1101/gr.103499.109. - DOI - PMC - PubMed
-
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

