RNA-seq: impact of RNA degradation on transcript quantification
- PMID: 24885439
- PMCID: PMC4071332
- DOI: 10.1186/1741-7007-12-42
RNA-seq: impact of RNA degradation on transcript quantification
Abstract
Background: The use of low quality RNA samples in whole-genome gene expression profiling remains controversial. It is unclear if transcript degradation in low quality RNA samples occurs uniformly, in which case the effects of degradation can be corrected via data normalization, or whether different transcripts are degraded at different rates, potentially biasing measurements of expression levels. This concern has rendered the use of low quality RNA samples in whole-genome expression profiling problematic. Yet, low quality samples (for example, samples collected in the course of fieldwork) are at times the sole means of addressing specific questions.
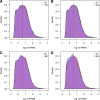
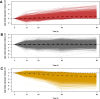
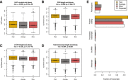
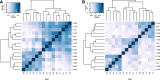
Results: We sought to quantify the impact of variation in RNA quality on estimates of gene expression levels based on RNA-seq data. To do so, we collected expression data from tissue samples that were allowed to decay for varying amounts of time prior to RNA extraction. The RNA samples we collected spanned the entire range of RNA Integrity Number (RIN) values (a metric commonly used to assess RNA quality). We observed widespread effects of RNA quality on measurements of gene expression levels, as well as a slight but significant loss of library complexity in more degraded samples.
Conclusions: While standard normalizations failed to account for the effects of degradation, we found that by explicitly controlling for the effects of RIN using a linear model framework we can correct for the majority of these effects. We conclude that in instances in which RIN and the effect of interest are not associated, this approach can help recover biologically meaningful signals in data from degraded RNA samples.
Figures

References
-
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, Amit I, Regev A. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;12:436–442. doi: 10.1038/nbt.1861. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

