Integrative analysis of the transcriptome profiles observed in type 1, type 2 and gestational diabetes mellitus reveals the role of inflammation
- PMID: 24885568
- PMCID: PMC4066312
- DOI: 10.1186/1755-8794-7-28
Integrative analysis of the transcriptome profiles observed in type 1, type 2 and gestational diabetes mellitus reveals the role of inflammation
Abstract
Background: Type 1 diabetes (T1D) is an autoimmune disease, while type 2 (T2D) and gestational diabetes (GDM) are considered metabolic disturbances. In a previous study evaluating the transcript profiling of peripheral mononuclear blood cells obtained from T1D, T2D and GDM patients we showed that the gene profile of T1D patients was closer to GDM than to T2D. To understand the influence of demographical, clinical, laboratory, pathogenetic and treatment features on the diabetes transcript profiling, we performed an analysis integrating these features with the gene expression profiles of the annotated genes included in databases containing information regarding GWAS and immune cell expression signatures.
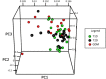
Methods: Samples from 56 (19 T1D, 20 T2D, and 17 GDM) patients were hybridized to whole genome one-color Agilent 4x44k microarrays. Non-informative genes were filtered by partitioning, and differentially expressed genes were obtained by rank product analysis. Functional analyses were carried out using the DAVID database, and module maps were constructed using the Genomica tool.
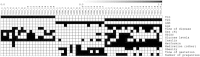
Results: The functional analyses were able to discriminate between T1D and GDM patients based on genes involved in inflammation. Module maps of differentially expressed genes revealed that modulated genes: i) exhibited transcription profiles typical of macrophage and dendritic cells; ii) had been previously associated with diabetic complications by association and by meta-analysis studies, and iii) were influenced by disease duration, obesity, number of gestations, glucose serum levels and the use of medications, such as metformin.
Conclusion: This is the first module map study to show the influence of epidemiological, clinical, laboratory, immunopathogenic and treatment features on the transcription profiles of T1D, T2D and GDM patients.
Figures





Similar articles
-
Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients.BMC Res Notes. 2013 Nov 26;6:491. doi: 10.1186/1756-0500-6-491. BMC Res Notes. 2013. PMID: 24279768 Free PMC article.
-
Identification of hub-methylated differentially expressed genes in patients with gestational diabetes mellitus by multi-omic WGCNA basing epigenome-wide and transcriptome-wide profiling.J Cell Biochem. 2020 Jun;121(5-6):3173-3184. doi: 10.1002/jcb.29584. Epub 2019 Dec 30. J Cell Biochem. 2020. PMID: 31886571
-
Comprehensive Analysis of Gene Expression Profiles and DNA Methylome reveals Oas1, Ppie, Polr2g as Pathogenic Target Genes of Gestational Diabetes Mellitus.Sci Rep. 2018 Nov 2;8(1):16244. doi: 10.1038/s41598-018-34292-z. Sci Rep. 2018. PMID: 30389953 Free PMC article.
-
Genetic Studies of Gestational Diabetes and Glucose Metabolism in Pregnancy.Curr Diab Rep. 2020 Nov 9;20(12):69. doi: 10.1007/s11892-020-01355-3. Curr Diab Rep. 2020. PMID: 33165656 Free PMC article. Review.
-
HNF1A:From Monogenic Diabetes to Type 2 Diabetes and Gestational Diabetes Mellitus.Front Endocrinol (Lausanne). 2022 Mar 1;13:829565. doi: 10.3389/fendo.2022.829565. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35299962 Free PMC article. Review.
Cited by
-
Pathophysiological impact of CXC and CX3CL1 chemokines in preeclampsia and gestational diabetes mellitus.Front Cell Dev Biol. 2023 Oct 19;11:1272536. doi: 10.3389/fcell.2023.1272536. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37928902 Free PMC article. Review.
-
First trimester secreted Frizzled-Related Protein 4 and other adipokine serum concentrations in women developing gestational diabetes mellitus.PLoS One. 2020 Nov 18;15(11):e0242423. doi: 10.1371/journal.pone.0242423. eCollection 2020. PLoS One. 2020. PMID: 33206702 Free PMC article.
-
Genetic determinants for gestational diabetes mellitus and related metabolic traits in Mexican women.PLoS One. 2015 May 14;10(5):e0126408. doi: 10.1371/journal.pone.0126408. eCollection 2015. PLoS One. 2015. PMID: 25973943 Free PMC article.
-
Cytotoxicity-Related Gene Expression and Chromatin Accessibility Define a Subset of CD4+ T Cells That Mark Progression to Type 1 Diabetes.Diabetes. 2022 Mar 1;71(3):566-577. doi: 10.2337/db21-0612. Diabetes. 2022. PMID: 35007320 Free PMC article.
-
Integrated analysis of mRNA and miRNA profiles revealed the role of miR-193 and miR-210 as potential regulatory biomarkers in different molecular subtypes of breast cancer.BMC Cancer. 2021 Jan 18;21(1):76. doi: 10.1186/s12885-020-07731-2. BMC Cancer. 2021. PMID: 33461524 Free PMC article.
References
-
- International Diabetes Federation. [ http://www.idf.org/diabetesatlas/]
-
- Lauenborg J, Hansen T, Jensen DM, Vestergaard H, Mølsted-Pedersen L, Hornnes P, Locht H, Pedersen O, Damm P. Increasing incidence of diabetes after gestational diabetes: a long-term follow-up in a Danish population. Diabetes Care. 2004;7:1194–1199. - PubMed
-
- Bellamy L, Casas J-P, Hingorani AD, Williams D. Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet. 2009;7:1773–1779. - PubMed
-
- Chentoufi AA, Binder NR, Berka N, Abunadi T, Polychronakos C. Advances in type I diabetes associated tolerance mechanisms. Scand J Immunol. 2008;7:1–11. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

