A variant of green fluorescent protein exclusively deposited to active intracellular inclusion bodies
- PMID: 24885571
- PMCID: PMC4049505
- DOI: 10.1186/1475-2859-13-68
A variant of green fluorescent protein exclusively deposited to active intracellular inclusion bodies
Abstract
Background: Inclusion bodies (IBs) were generally considered to be inactive protein deposits and did not hold any attractive values in biotechnological applications. Recently, some IBs of recombinant proteins were confirmed to show their functional properties such as enzyme activities, fluorescence, etc. Such biologically active IBs are not commonly formed, but they have great potentials in the fields of biocatalysis, material science and nanotechnology.
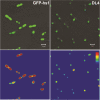
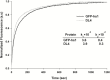
Results: In this study, we characterized the IBs of DL4, a deletion variant of green fluorescent protein which forms active intracellular aggregates. The DL4 proteins expressed in Escherichia coli were exclusively deposited to IBs, and the IBs were estimated to be mostly composed of active proteins. The spectral properties and quantum yield of the DL4 variant in the active IBs were almost same with those of its native protein. Refolding and stability studies revealed that the deletion mutation in DL4 didn't affect the folding efficiency of the protein, but destabilized its structure. Analyses specific for amyloid-like structures informed that the inner architecture of DL4 IBs might be amorphous rather than well-organized. The diameter of fluorescent DL4 IBs could be decreased up to 100-200 nm by reducing the expression time of the protein in vivo.
Conclusions: To our knowledge, DL4 is the first GFP variant that folds correctly but aggregates exclusively in vivo without any self-aggregating/assembling tags. The fluorescent DL4 IBs have potentials to be used as fluorescent biomaterials. This study also suggests that biologically active IBs can be achieved through engineering a target protein itself.
Figures






Similar articles
-
Studies on the Structure and Properties of Membrane Phospholipase A1 Inclusion Bodies Formed at Low Growth Temperatures Using GFP Fusion Strategy.Molecules. 2021 Jun 28;26(13):3936. doi: 10.3390/molecules26133936. Molecules. 2021. PMID: 34203222 Free PMC article.
-
Exploring the use of leucine zippers for the generation of a new class of inclusion bodies for pharma and biotechnological applications.Microb Cell Fact. 2020 Sep 4;19(1):175. doi: 10.1186/s12934-020-01425-x. Microb Cell Fact. 2020. PMID: 32887587 Free PMC article.
-
Active inclusion bodies of acid phosphatase PhoC: aggregation induced by GFP fusion and activities modulated by linker flexibility.Microb Cell Fact. 2013 Mar 14;12:25. doi: 10.1186/1475-2859-12-25. Microb Cell Fact. 2013. PMID: 23497261 Free PMC article.
-
Active protein aggregates produced in Escherichia coli.Int J Mol Sci. 2011;12(11):8275-87. doi: 10.3390/ijms12118275. Epub 2011 Nov 22. Int J Mol Sci. 2011. PMID: 22174663 Free PMC article. Review.
-
Reassessment of inclusion body-based production as a versatile opportunity for difficult-to-express recombinant proteins.Crit Rev Biotechnol. 2018 Aug;38(5):729-744. doi: 10.1080/07388551.2017.1398134. Epub 2017 Nov 10. Crit Rev Biotechnol. 2018. PMID: 29124949 Review.
Cited by
-
Formation of Biomolecular Condensates in Bacteria by Tuning Protein Electrostatics.ACS Cent Sci. 2020 Dec 23;6(12):2301-2310. doi: 10.1021/acscentsci.0c01146. Epub 2020 Nov 12. ACS Cent Sci. 2020. PMID: 33376791 Free PMC article.
-
Studies on the Structure and Properties of Membrane Phospholipase A1 Inclusion Bodies Formed at Low Growth Temperatures Using GFP Fusion Strategy.Molecules. 2021 Jun 28;26(13):3936. doi: 10.3390/molecules26133936. Molecules. 2021. PMID: 34203222 Free PMC article.
-
Soluble Expression of hFGF19 without Fusion Protein through Synonymous Codon Substitutions and DsbC Co-Expression in E. coli.Microorganisms. 2020 Dec 7;8(12):1942. doi: 10.3390/microorganisms8121942. Microorganisms. 2020. PMID: 33297586 Free PMC article.
-
Formation of active inclusion bodies induced by hydrophobic self-assembling peptide GFIL8.Microb Cell Fact. 2015 Jun 16;14:88. doi: 10.1186/s12934-015-0270-0. Microb Cell Fact. 2015. PMID: 26077447 Free PMC article.
-
Plasmids for Controlled and Tunable High-Level Expression in E. coli.Appl Environ Microbiol. 2022 Nov 22;88(22):e0093922. doi: 10.1128/aem.00939-22. Epub 2022 Nov 7. Appl Environ Microbiol. 2022. PMID: 36342148 Free PMC article.
References
-
- Fink AL. Chaperone-mediated protein folding. Physiol Rev. 1999;79:425–449. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

