Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq
- PMID: 24885638
- PMCID: PMC4036795
- DOI: 10.1186/1471-2229-14-120
Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq
Abstract
Background: The availability of draft crop plant genomes allows the prediction of the full complement of genes that encode NB-LRR resistance gene homologs, enabling a more targeted breeding for disease resistance. Recently, we developed the RenSeq method to reannotate the full NB-LRR gene complement in potato and to identify novel sequences that were not picked up by the automated gene prediction software. Here, we established RenSeq on the reference genome of tomato (Solanum lycopersicum) Heinz 1706, using 260 previously identified NB-LRR genes in an updated Solanaceae RenSeq bait library.
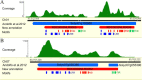
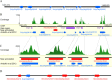
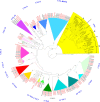
Result: Using 250-bp MiSeq reads after RenSeq on genomic DNA of Heinz 1706, we identified 105 novel NB-LRR sequences. Reannotation included the splitting of gene models, combination of partial genes to a longer sequence and closing of assembly gaps. Within the draft S. pimpinellifolium LA1589 genome, RenSeq enabled the annotation of 355 NB-LRR genes. The majority of these are however fragmented, with 5'- and 3'-end located on the edges of separate contigs. Phylogenetic analyses show a high conservation of all NB-LRR classes between Heinz 1706, LA1589 and the potato clone DM, suggesting that all sub-families were already present in the last common ancestor. A phylogenetic comparison to the Arabidopsis thaliana NB-LRR complement verifies the high conservation of the more ancient CCRPW8-type NB-LRRs. Use of RenSeq on cDNA from uninfected and late blight-infected tomato leaves allows the avoidance of sequence analysis of non-expressed paralogues.
Conclusion: RenSeq is a promising method to facilitate analysis of plant resistance gene complements. The reannotated tomato NB-LRR complements, phylogenetic relationships and chromosomal locations provided in this paper will provide breeders and scientists with a useful tool to identify novel disease resistance traits. cDNA RenSeq enables for the first time next-gen sequencing approaches targeted to this very low-expressed gene family without the need for normalization.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

