GAP-Seq: a method for identification of DNA palindromes
- PMID: 24885769
- PMCID: PMC4057610
- DOI: 10.1186/1471-2164-15-394
GAP-Seq: a method for identification of DNA palindromes
Abstract
Background: Closely spaced long inverted repeats, also known as DNA palindromes, can undergo intrastrand annealing to form DNA hairpins. The ability to form these hairpins results in genome instability, difficulties in maintaining clones in Escherichia coli and major problems for most DNA sequencing approaches. Because of their role in genomic instability and gene amplification in some human cancers, it is important to develop systematic approaches to detect and characterize DNA palindromes.
Results: We developed a new protocol to identify palindromes that couples the S1 nuclease treated Cot0 DNA (GAPF) with high-throughput sequencing (GAP-Seq). Unlike earlier protocols, it does not involve restriction enzymatic digestion prior to DNA snap-back thereby preserving longer DNA sequences. It also indicates the location of the novel junction, which can then be recovered. Using MCF-7 breast cancer cell line as the proof-of-principle analysis, we have identified 35 palindrome candidates and physically characterized the top 5 candidates and their junctions. Because this protocol eliminates many of the false positives that plague earlier techniques, we have improved palindrome identification.
Conclusions: The GAP-Seq approach underscores the importance of developing new tools for identifying and characterizing palindromes, and provides a new strategy to systematically assess palindromes in genomes. It will be useful for studying human cancers and other diseases associated with palindromes.
Figures

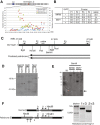
 . 3: Clustering two or more contigs with a base read ratio >1.5 that are within 7.5 kb of each other to make a joined contig. 4: Determining Rank “R” that is the sum of all read lengths in the joined contig divided by the length of the joined contig minus the masked regions
. 3: Clustering two or more contigs with a base read ratio >1.5 that are within 7.5 kb of each other to make a joined contig. 4: Determining Rank “R” that is the sum of all read lengths in the joined contig divided by the length of the joined contig minus the masked regions  .
.


Similar articles
-
Genome-Wide Analysis of Palindrome Formation with Next-Generation Sequencing (GAPF-Seq) and a Bioinformatics Pipeline for Assessing De Novo Palindromes in Cancer Genomes.Methods Mol Biol. 2023;2660:13-22. doi: 10.1007/978-1-0716-3163-8_2. Methods Mol Biol. 2023. PMID: 37191787
-
Palindromes in DNA-A Risk for Genome Stability and Implications in Cancer.Int J Mol Sci. 2021 Mar 11;22(6):2840. doi: 10.3390/ijms22062840. Int J Mol Sci. 2021. PMID: 33799581 Free PMC article. Review.
-
Large DNA palindromes as a common form of structural chromosome aberrations in human cancers.Hum Cell. 2006 Feb;19(1):17-23. doi: 10.1111/j.1749-0774.2005.00003.x. Hum Cell. 2006. PMID: 16643603
-
Myc oncogene-induced genomic instability: DNA palindromes in bursal lymphomagenesis.PLoS Genet. 2008 Jul 18;4(7):e1000132. doi: 10.1371/journal.pgen.1000132. PLoS Genet. 2008. PMID: 18636108 Free PMC article.
-
Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair.Bioessays. 1994 Dec;16(12):893-900. doi: 10.1002/bies.950161207. Bioessays. 1994. PMID: 7840768 Review.
Cited by
-
Coding palindromes in mitochondrial genes of Nematomorpha.Nucleic Acids Res. 2019 Jul 26;47(13):6858-6870. doi: 10.1093/nar/gkz517. Nucleic Acids Res. 2019. PMID: 31194871 Free PMC article.
-
Genome-Wide Analysis of Palindrome Formation with Next-Generation Sequencing (GAPF-Seq) and a Bioinformatics Pipeline for Assessing De Novo Palindromes in Cancer Genomes.Methods Mol Biol. 2023;2660:13-22. doi: 10.1007/978-1-0716-3163-8_2. Methods Mol Biol. 2023. PMID: 37191787
-
Origin-Dependent Inverted-Repeat Amplification: Tests of a Model for Inverted DNA Amplification.PLoS Genet. 2015 Dec 23;11(12):e1005699. doi: 10.1371/journal.pgen.1005699. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26700858 Free PMC article.
-
Palindromes in DNA-A Risk for Genome Stability and Implications in Cancer.Int J Mol Sci. 2021 Mar 11;22(6):2840. doi: 10.3390/ijms22062840. Int J Mol Sci. 2021. PMID: 33799581 Free PMC article. Review.
-
Double insertion of transposable elements provides a substrate for the evolution of satellite DNA.Genome Res. 2018 May;28(5):714-725. doi: 10.1101/gr.231472.117. Epub 2018 Mar 27. Genome Res. 2018. PMID: 29588362 Free PMC article.
References
-
- Inagaki H, Ohye T, Kogo H, Kato T, Bolor H, Taniguchi M, Shaikh TH, Emanuel BS, Kurahashi H. Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans. Genome Res. 2009;19(2):191–198. doi: 10.1101/gr.079244.108. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- SRA/SRA064847
- SRA/SRA065361
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

