QGRS-Conserve: a computational method for discovering evolutionarily conserved G-quadruplex motifs
- PMID: 24885782
- PMCID: PMC4017754
- DOI: 10.1186/1479-7364-8-8
QGRS-Conserve: a computational method for discovering evolutionarily conserved G-quadruplex motifs
Abstract
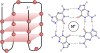
Background: Nucleic acids containing guanine tracts can form quadruplex structures via non-Watson-Crick base pairing. Formation of G-quadruplexes is associated with the regulation of important biological functions such as transcription, genetic instability, DNA repair, DNA replication, epigenetic mechanisms, regulation of translation, and alternative splicing. G-quadruplexes play important roles in human diseases and are being considered as targets for a variety of therapies. Identification of functional G-quadruplexes and the study of their overall distribution in genomes and transcriptomes is an important pursuit. Traditional computational methods map sequence motifs capable of forming G-quadruplexes but have difficulty in distinguishing motifs that occur by chance from ones which fold into G-quadruplexes.
Results: We present Quadruplex forming 'G'-rich sequences (QGRS)-Conserve, a computational method for calculating motif conservation across exomes and supports filtering to provide researchers with more precise methods of studying G-quadruplex distribution patterns. Our method quantitatively evaluates conservation between quadruplexes found in homologous nucleotide sequences based on several motif structural characteristics. QGRS-Conserve also efficiently manages overlapping G-quadruplex sequences such that the resulting datasets can be analyzed effectively.
Conclusions: We have applied QGRS-Conserve to identify a large number of G-quadruplex motifs in the human exome conserved across several mammalian and non-mammalian species. We have successfully identified multiple homologs of many previously published G-quadruplexes that play post-transcriptional regulatory roles in human genes. Preliminary large-scale analysis identified many homologous G-quadruplexes in the 5'- and 3'-untranslated regions of mammalian species. An expectedly smaller set of G-quadruplex motifs was found to be conserved across larger phylogenetic distances. QGRS-Conserve provides means to build datasets that can be filtered and categorized in a variety of biological dimensions for more targeted studies in order to better understand the roles that G-quadruplexes play.
Figures


Similar articles
-
QGRS-H Predictor: a web server for predicting homologous quadruplex forming G-rich sequence motifs in nucleotide sequences.Nucleic Acids Res. 2012 Jul;40(Web Server issue):W96-W103. doi: 10.1093/nar/gks422. Epub 2012 May 10. Nucleic Acids Res. 2012. PMID: 22576365 Free PMC article.
-
QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W676-82. doi: 10.1093/nar/gkl253. Nucleic Acids Res. 2006. PMID: 16845096 Free PMC article.
-
Role of conserved cis-regulatory elements in the post-transcriptional regulation of the human MECP2 gene involved in autism.Hum Genomics. 2013 Sep 16;7(1):19. doi: 10.1186/1479-7364-7-19. Hum Genomics. 2013. PMID: 24040966 Free PMC article.
-
Beyond G-Quadruplexes-The Effect of Junction with Additional Structural Motifs on Aptamers Properties.Int J Mol Sci. 2021 Sep 14;22(18):9948. doi: 10.3390/ijms22189948. Int J Mol Sci. 2021. PMID: 34576112 Free PMC article. Review.
-
An Updated Focus on Quadruplex Structures as Potential Therapeutic Targets in Cancer.Int J Mol Sci. 2020 Nov 24;21(23):8900. doi: 10.3390/ijms21238900. Int J Mol Sci. 2020. PMID: 33255335 Free PMC article. Review.
Cited by
-
Detection of G-type density in promoter sequence of colon cancer oncogenes and tumor suppressor genes.Bioinformation. 2015 Jun 30;11(6):290-5. doi: 10.6026/97320630011290. eCollection 2015. Bioinformation. 2015. PMID: 26229289 Free PMC article.
-
G-quadruplex prediction in E. coli genome reveals a conserved putative G-quadruplex-Hairpin-Duplex switch.Nucleic Acids Res. 2016 Nov 2;44(19):9083-9095. doi: 10.1093/nar/gkw769. Epub 2016 Sep 4. Nucleic Acids Res. 2016. PMID: 27596596 Free PMC article.
-
Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants.Sci Rep. 2016 Jun 21;6:28211. doi: 10.1038/srep28211. Sci Rep. 2016. PMID: 27324275 Free PMC article.
-
G4Boost: a machine learning-based tool for quadruplex identification and stability prediction.BMC Bioinformatics. 2022 Jun 18;23(1):240. doi: 10.1186/s12859-022-04782-z. BMC Bioinformatics. 2022. PMID: 35717172 Free PMC article.
-
Conjunction of potential G-quadruplex and adjacent cis-elements in the 5' UTR of hepatocyte nuclear factor 4-alpha strongly inhibit protein expression.Sci Rep. 2017 Dec 12;7(1):17444. doi: 10.1038/s41598-017-17629-y. Sci Rep. 2017. PMID: 29234104 Free PMC article.
References
-
- Simonsson T. G-quadruplex DNA structures–variations on a theme. Biol Chem. 2001;382:621–628. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

