Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana
- PMID: 24885787
- PMCID: PMC4081654
- DOI: 10.1186/1471-2229-14-141
Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana
Abstract
Background: Pre-exposing plants to diverse abiotic stresses may alter their physiological and transcriptional responses to a subsequent stress, suggesting a form of "stress memory". Arabidopsis thaliana plants that have experienced multiple exposures to dehydration stress display transcriptional behavior suggesting "memory" from an earlier stress. Genes that respond to a first stress by up-regulating or down-regulating their transcription but in a subsequent stress provide a significantly different response define the 'memory genes' category. Genes responding similarly to each stress form the 'non-memory' category. It is unknown whether such memory responses exists in other Angiosperm lineages and whether memory is an evolutionarily conserved response to repeated dehydration stresses.
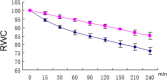
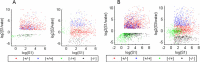
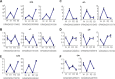
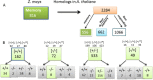
Results: Here, we determine the transcriptional responses of maize (Zea mays L.) plants that have experienced repeated exposures to dehydration stress in comparison with plants encountering the stress for the first time. Four distinct transcription memory response patterns similar to those displayed by A. thaliana were revealed. The most important contribution is the evidence that monocot and eudicot plants, two lineages that have diverged 140 to 200 M years ago, display similar abilities to 'remember' a dehydration stress and to modify their transcriptional responses, accordingly. The highly sensitive RNA-Seq analyses allowed to identify genes that function similarly in the two lineages, as well as genes that function in species-specific ways. Memory transcription patterns indicate that the transcriptional behavior of responding genes under repeated stresses is different from the behavior during an initial dehydration stress, suggesting that stress memory is a complex phenotype resulting from coordinated responses of multiple signaling pathways.
Conclusions: Structurally related genes displaying the same memory responses in the two species would suggest conservation of the genes' memory during the evolution of plants' dehydration stress response systems. On the other hand, divergent transcription memory responses by genes encoding similar functions would suggest occurrence of species-specific memory responses. The results provide novel insights into our current knowledge of how plants respond to multiple dehydration stresses, as compared to a single exposure, and may serve as a reference platform to study the functions of memory genes in adaptive responses to water deficit in monocot and eudicot plants.
Figures


Similar articles
-
Four distinct types of dehydration stress memory genes in Arabidopsis thaliana.BMC Plant Biol. 2013 Dec 30;13:229. doi: 10.1186/1471-2229-13-229. BMC Plant Biol. 2013. PMID: 24377444 Free PMC article.
-
Different gene-specific mechanisms determine the 'revised-response' memory transcription patterns of a subset of A. thaliana dehydration stress responding genes.Nucleic Acids Res. 2014 May;42(9):5556-66. doi: 10.1093/nar/gku220. Epub 2014 Apr 17. Nucleic Acids Res. 2014. PMID: 24744238 Free PMC article.
-
ABA signaling is necessary but not sufficient for RD29B transcriptional memory during successive dehydration stresses in Arabidopsis thaliana.Plant J. 2014 Jul;79(1):150-61. doi: 10.1111/tpj.12548. Epub 2014 Jun 17. Plant J. 2014. PMID: 24805058
-
Transcriptional 'memory' of a stress: transient chromatin and memory (epigenetic) marks at stress-response genes.Plant J. 2015 Jul;83(1):149-59. doi: 10.1111/tpj.12832. Epub 2015 Apr 15. Plant J. 2015. PMID: 25788029 Review.
-
Functional genomics in plant abiotic stress responses and tolerance: From gene discovery to complex regulatory networks and their application in breeding.Proc Jpn Acad Ser B Phys Biol Sci. 2022;98(8):470-492. doi: 10.2183/pjab.98.024. Proc Jpn Acad Ser B Phys Biol Sci. 2022. PMID: 36216536 Free PMC article. Review.
Cited by
-
Transcriptional and physiological data reveal the dehydration memory behavior in switchgrass (Panicum virgatum L.).Biotechnol Biofuels. 2018 Apr 2;11:91. doi: 10.1186/s13068-018-1088-x. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29619087 Free PMC article.
-
Memory or acclimation of water stress in pea rely on root system's plasticity and plant's ionome modulation.Front Plant Sci. 2023 Jan 25;13:1089720. doi: 10.3389/fpls.2022.1089720. eCollection 2022. Front Plant Sci. 2023. PMID: 36762182 Free PMC article.
-
Drought Tolerance Strategies and Autophagy in Resilient Wheat Genotypes.Cells. 2022 May 27;11(11):1765. doi: 10.3390/cells11111765. Cells. 2022. PMID: 35681460 Free PMC article.
-
Long non-coding RNAs of switchgrass (Panicum virgatum L.) in multiple dehydration stresses.BMC Plant Biol. 2018 May 4;18(1):79. doi: 10.1186/s12870-018-1288-3. BMC Plant Biol. 2018. PMID: 29728055 Free PMC article.
-
Drought Stress Memory at the Plant Cycle Level: A Review.Plants (Basel). 2021 Sep 10;10(9):1873. doi: 10.3390/plants10091873. Plants (Basel). 2021. PMID: 34579406 Free PMC article. Review.
References
-
- Levitt J. Responses of plants to environmental stresses. 2d. New York: Academic Press; 1980.
-
- Bruce TJA, Matthes MC, Napier JA, Pickett JA. Stressful “memories” of plants: Evidence and possible mechanisms. Plant Sci. 2007;173(6):603–608. doi: 10.1016/j.plantsci.2007.09.002. - DOI
-
- Conrath U, Beckers GJM, Flors V, Garcia-Agustin P, Jakab G, Mauch F, Newman MA, Pieterse CMJ, Poinssot B, Pozo MJ, Pugin A, Schaffrath U, Ton J, Wendehenne D, Zimmerli L, Mauch-Mani B. Priming: Getting ready for battle. Mol Plant Microbe Interact. 2006;19(10):1062–1071. doi: 10.1094/MPMI-19-1062. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

