Multiple modes of proepicardial cell migration require heartbeat
- PMID: 24885804
- PMCID: PMC4048602
- DOI: 10.1186/1471-213X-14-18
Multiple modes of proepicardial cell migration require heartbeat
Abstract
Background: The outermost layer of the vertebrate heart, the epicardium, forms from a cluster of progenitor cells termed the proepicardium (PE). PE cells migrate onto the myocardium to give rise to the epicardium. Impaired epicardial development has been associated with defects in valve development, cardiomyocyte proliferation and alignment, cardiac conduction system maturation and adult heart regeneration. Zebrafish are an excellent model for studying cardiac development and regeneration; however, little is known about how the zebrafish epicardium forms.
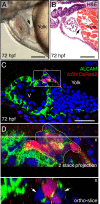
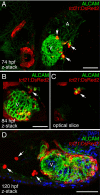
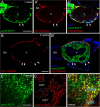
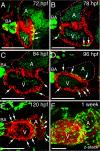
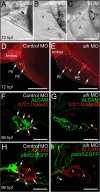
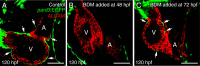
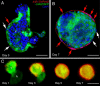
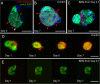
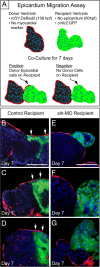
Results: We report that PE migration occurs through multiple mechanisms and that the zebrafish epicardium is composed of a heterogeneous population of cells. Heterogeneity is first observed within the PE and persists through epicardium formation. Using in vivo imaging, histology and confocal microscopy, we show that PE cells migrate through a cellular bridge that forms between the pericardial mesothelium and the heart. We also observed the formation of PE aggregates on the pericardial surface, which were released into the pericardial cavity. It was previously reported that heartbeat-induced pericardiac fluid advections are necessary for PE cluster formation and subsequent epicardium development. We manipulated heartbeat genetically and pharmacologically and found that PE clusters clearly form in the absence of heartbeat. However, when heartbeat was inhibited the PE failed to migrate to the myocardium and the epicardium did not form. We isolated and cultured hearts with only a few epicardial progenitor cells and found a complete epicardial layer formed. However, pharmacologically inhibiting contraction in culture prevented epicardium formation. Furthermore, we isolated control and silent heart (sih) morpholino (MO) injected hearts prior to epicardium formation (60 hpf) and co-cultured these hearts with "donor" hearts that had an epicardium forming (108 hpf). Epicardial cells from donor hearts migrated on to control but not sih MO injected hearts.
Conclusions: Epicardial cells stem from a heterogeneous population of progenitors, suggesting that the progenitors in the PE have distinct identities. PE cells attach to the heart via a cellular bridge and free-floating cell clusters. Pericardiac fluid advections are not necessary for the development of the PE cluster, however heartbeat is required for epicardium formation. Epicardium formation can occur in culture without normal hydrodynamic and hemodynamic forces, but not without contraction.
Figures
References
-
- Vincent SD, Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol. 2010;90:1–41. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

