Differential regulation of the α-globin locus by Krüppel-like Factor 3 in erythroid and non-erythroid cells
- PMID: 24885809
- PMCID: PMC4033687
- DOI: 10.1186/1471-2199-15-8
Differential regulation of the α-globin locus by Krüppel-like Factor 3 in erythroid and non-erythroid cells
Abstract
Background: Krüppel-like Factor 3 (KLF3) is a broadly expressed zinc-finger transcriptional repressor with diverse biological roles. During erythropoiesis, KLF3 acts as a feedback repressor of a set of genes that are activated by Krüppel-like Factor 1 (KLF1). Noting that KLF1 binds α-globin gene regulatory sequences during erythroid maturation, we sought to determine whether KLF3 also interacts with the α-globin locus to regulate transcription.
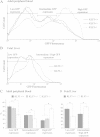
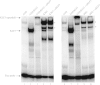
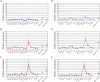
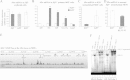
Results: We found that expression of a human transgenic α-globin reporter gene is markedly up-regulated in fetal and adult erythroid cells of Klf3-/- mice. Inspection of the mouse and human α-globin promoters revealed a number of canonical KLF-binding sites, and indeed, KLF3 was shown to bind to these regions both in vitro and in vivo. Despite these observations, we did not detect an increase in endogenous murine α-globin expression in Klf3-/- erythroid tissue. However, examination of murine embryonic fibroblasts lacking KLF3 revealed significant de-repression of α-globin gene expression. This suggests that KLF3 may contribute to the silencing of the α-globin locus in non-erythroid tissue. Moreover, ChIP-Seq analysis of murine fibroblasts demonstrated that across the locus, KLF3 does not occupy the promoter regions of the α-globin genes in these cells, but rather, binds to upstream, DNase hypersensitive regulatory regions.
Conclusions: These findings reveal that the occupancy profile of KLF3 at the α-globin locus differs in erythroid and non-erythroid cells. In erythroid cells, KLF3 primarily binds to the promoters of the adult α-globin genes, but appears dispensable for normal transcriptional regulation. In non-erythroid cells, KLF3 distinctly binds to the HS-12 and HS-26 elements and plays a non-redundant, albeit modest, role in the silencing of α-globin expression.
Figures


References
-
- Pearson RC, Funnell AP, Crossley M. The mammalian zinc finger transcription factor Kruppel-like factor 3 (KLF3/BKLF) IUBMB life. 2011;63(2):86–93. - PubMed
-
- Funnell AP, Norton LJ, Mak KS, Burdach J, Artuz CM, Twine NA, Wilkins MR, Power CA, Hung TT, Perdomo J, Koh P, Bell-Anderson KS, Orkin SH, Fraser ST, Perkins AC, Pearson RC, Crossley M. The CACCC-binding protein KLF3/BKLF represses a subset of KLF1/EKLF target genes and is required for proper erythroid maturation in vivo. Mol Cell Biol. 2012;32(16):3281–3292. doi: 10.1128/MCB.00173-12. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

