Stability of gene expression and epigenetic profiles highlights the utility of patient-derived paediatric acute lymphoblastic leukaemia xenografts for investigating molecular mechanisms of drug resistance
- PMID: 24885906
- PMCID: PMC4057609
- DOI: 10.1186/1471-2164-15-416
Stability of gene expression and epigenetic profiles highlights the utility of patient-derived paediatric acute lymphoblastic leukaemia xenografts for investigating molecular mechanisms of drug resistance
Abstract
Background: Patient-derived tumour xenografts are an attractive model for preclinical testing of anti-cancer drugs. Insights into tumour biology and biomarkers predictive of responses to chemotherapeutic drugs can also be gained from investigating xenograft models. As a first step towards examining the equivalence of epigenetic profiles between xenografts and primary tumours in paediatric leukaemia, we performed genome-scale DNA methylation and gene expression profiling on a panel of 10 paediatric B-cell precursor acute lymphoblastic leukaemia (BCP-ALL) tumours that were stratified by prednisolone response.
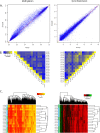
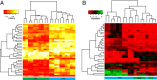
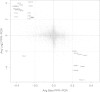
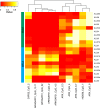
Results: We found high correlations in DNA methylation and gene expression profiles between matching primary and xenograft tumour samples with Pearson's correlation coefficients ranging between 0.85 and 0.98. In order to demonstrate the potential utility of epigenetic analyses in BCP-ALL xenografts, we identified DNA methylation biomarkers that correlated with prednisolone responsiveness of the original tumour samples. Differential methylation of CAPS2, ARHGAP21, ARX and HOXB6 were confirmed by locus specific analysis. We identified 20 genes showing an inverse relationship between DNA methylation and gene expression in association with prednisolone response. Pathway analysis of these genes implicated apoptosis, cell signalling and cell structure networks in prednisolone responsiveness.
Conclusions: The findings of this study confirm the stability of epigenetic and gene expression profiles of paediatric BCP-ALL propagated in mouse xenograft models. Further, our preliminary investigation of prednisolone sensitivity highlights the utility of mouse xenograft models for preclinical development of novel drug regimens with parallel investigation of underlying gene expression and epigenetic responses associated with novel drug responses.
Figures
Similar articles
-
p53-independent epigenetic repression of the p21(WAF1) gene in T-cell acute lymphoblastic leukemia.J Biol Chem. 2011 Oct 28;286(43):37639-50. doi: 10.1074/jbc.M111.272336. Epub 2011 Sep 7. J Biol Chem. 2011. PMID: 21903579 Free PMC article.
-
Apoptosis Induced by Prednisolone Occurs without Altering the Promoter Methylation of BAX and BCL-2 Genes in Acute Lymphoblastic Leukemia Cells CCRF-CEM.Asian Pac J Cancer Prev. 2020 Feb 1;21(2):523-529. doi: 10.31557/APJCP.2020.21.2.523. Asian Pac J Cancer Prev. 2020. PMID: 32102534 Free PMC article.
-
Genome-wide methylation profiling of ovarian cancer patient-derived xenografts treated with the demethylating agent decitabine identifies novel epigenetically regulated genes and pathways.Genome Med. 2016 Oct 20;8(1):107. doi: 10.1186/s13073-016-0361-5. Genome Med. 2016. PMID: 27765068 Free PMC article.
-
Epigenetic Control of Infant B Cell Precursor Acute Lymphoblastic Leukemia.Int J Mol Sci. 2021 Mar 18;22(6):3127. doi: 10.3390/ijms22063127. Int J Mol Sci. 2021. PMID: 33803872 Free PMC article. Review.
-
Can we use epigenetics to prime chemoresistant lymphomas?Hematology Am Soc Hematol Educ Program. 2020 Dec 4;2020(1):85-94. doi: 10.1182/hematology.2020000092. Hematology Am Soc Hematol Educ Program. 2020. PMID: 33275728 Free PMC article. Review.
Cited by
-
Establishment of a patient-derived orthotopic osteosarcoma mouse model.J Transl Med. 2015 Apr 30;13:136. doi: 10.1186/s12967-015-0497-x. J Transl Med. 2015. PMID: 25926029 Free PMC article.
-
Antileukemic Efficacy of Continuous vs Discontinuous Dexamethasone in Murine Models of Acute Lymphoblastic Leukemia.PLoS One. 2015 Aug 7;10(8):e0135134. doi: 10.1371/journal.pone.0135134. eCollection 2015. PLoS One. 2015. PMID: 26252865 Free PMC article.
-
Failure of a patient-derived xenograft for brain tumor model prepared by implantation of tissue fragments.Cancer Cell Int. 2016 Jun 10;16:43. doi: 10.1186/s12935-016-0319-0. eCollection 2016. Cancer Cell Int. 2016. PMID: 27293382 Free PMC article.
-
Protocol for the avatar acceptability study: a multiperspective cross-sectional study evaluating the acceptability of using patient-derived xenografts to guide personalised cancer care in Australia and New Zealand.BMJ Open. 2018 Aug 8;8(8):e024064. doi: 10.1136/bmjopen-2018-024064. BMJ Open. 2018. PMID: 30093523 Free PMC article.
-
ARHGAP10, downregulated in ovarian cancer, suppresses tumorigenicity of ovarian cancer cells.Cell Death Dis. 2016 Mar 24;7(3):e2157. doi: 10.1038/cddis.2015.401. Cell Death Dis. 2016. PMID: 27010858 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

