Validation of housekeeping gene and impact on normalized gene expression in clear cell renal cell carcinoma: critical reassessment of YBX3/ZONAB/CSDA expression
- PMID: 24885929
- PMCID: PMC4045873
- DOI: 10.1186/1471-2199-15-9
Validation of housekeeping gene and impact on normalized gene expression in clear cell renal cell carcinoma: critical reassessment of YBX3/ZONAB/CSDA expression
Abstract
Background: YBX3/ZONAB/CSDA is an epithelial-specific transcription factor acting in the density-based switch between proliferation and differentiation. Our laboratory reported overexpression of YBX3 in clear cell renal cell arcinoma (ccRCC), as part of a wide study of YBX3 regulation in vitro and in vivo. The preliminary data was limited to 5 cases, of which only 3 could be compared to paired normal tissue, and beta-Actin was used as sole reference to normalize gene expression. We thus decided to re-evaluate YBX3 expression by real-time-PCR in a larger panel of ccRCC samples, and their paired healthy tissue, with special attention on experimental biases such as inter-individual variations, primer specificity, and reference gene for normalization.
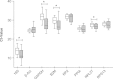
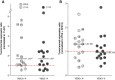
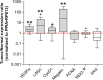
Results: Gene expression was measured by RT-qPCR in 16 ccRCC samples, each compared to corresponding healthy tissue to minimize inter-individual variations. Eight potential housekeeping genes were evaluated for expression level and stability among the 16-paired samples. Among tested housekeeping genes, PPIA and RPS13, especially in combination, proved best suitable to normalize gene expression in ccRCC tissues as compared to classical reference genes such as beta-Actin, GAPDH, 18S or B2M. Using this pair as reference, YBX3 expression level among a collection of 16 ccRCC tumors was not significantly increased as compared to normal adjacent tissues. However, stratification according to Fuhrman grade disclosed higher YBX3 expression levels in low-grade tumors and lower in high-grade tumors. Immunoperoxidase confirmed homogeneous nuclear staining for YBX3 in low-grade but revealed nuclear heterogeneity in high-grade tumors.
Conclusions: This paper underlines that special attention to reference gene products in the design of real-time PCR analysis of tumoral tissue is crucial to avoid misleading conclusions. Furthermore, we found that global YBX3/ZONAB/CSDA mRNA expression level may be considered within a "signature" of RCC grading.
Figures

Similar articles
-
Identification of RNA-binding protein YBX3 as an oncogene in clear cell renal cell carcinoma.Funct Integr Genomics. 2023 Jul 7;23(3):225. doi: 10.1007/s10142-023-01145-6. Funct Integr Genomics. 2023. PMID: 37418046 Free PMC article.
-
ZONAB promotes proliferation and represses differentiation of proximal tubule epithelial cells.J Am Soc Nephrol. 2010 Mar;21(3):478-88. doi: 10.1681/ASN.2009070698. Epub 2010 Feb 4. J Am Soc Nephrol. 2010. PMID: 20133480 Free PMC article.
-
In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR.BMC Mol Biol. 2007 Jun 8;8:47. doi: 10.1186/1471-2199-8-47. BMC Mol Biol. 2007. PMID: 17559644 Free PMC article.
-
Resolving the current controversy of use and reuse of housekeeping proteins in ageing research: Focus on saving people's tax dollars.Ageing Res Rev. 2024 Sep;100:102437. doi: 10.1016/j.arr.2024.102437. Epub 2024 Jul 25. Ageing Res Rev. 2024. PMID: 39067773 Review.
-
Un-biased housekeeping gene panel selection for high-validity gene expression analysis.Sci Rep. 2022 Jul 19;12(1):12324. doi: 10.1038/s41598-022-15989-8. Sci Rep. 2022. PMID: 35853974 Free PMC article.
Cited by
-
YBX3 Mediates the Metastasis of Nasopharyngeal Carcinoma via PI3K/AKT Signaling.Front Oncol. 2021 Mar 17;11:617621. doi: 10.3389/fonc.2021.617621. eCollection 2021. Front Oncol. 2021. PMID: 33816248 Free PMC article.
-
Ppia is the most stable housekeeping gene for qRT-PCR normalization in kidneys of three Pkd1-deficient mouse models.Sci Rep. 2021 Oct 5;11(1):19798. doi: 10.1038/s41598-021-99366-x. Sci Rep. 2021. PMID: 34611276 Free PMC article.
-
Adaptations of lipid metabolism in low-grade clear cell renal cell carcinoma are linked to cholesteryl ester accumulation.Sci Rep. 2025 Jul 9;15(1):24762. doi: 10.1038/s41598-025-09664-x. Sci Rep. 2025. PMID: 40634478 Free PMC article.
-
HOTAIRM1 lncRNA is downregulated in clear cell renal cell carcinoma and inhibits the hypoxia pathway.Cancer Lett. 2020 Mar 1;472:50-58. doi: 10.1016/j.canlet.2019.12.022. Epub 2019 Dec 17. Cancer Lett. 2020. PMID: 31862408 Free PMC article.
-
Automated identification of reference genes based on RNA-seq data.Biomed Eng Online. 2017 Aug 18;16(Suppl 1):65. doi: 10.1186/s12938-017-0356-5. Biomed Eng Online. 2017. PMID: 28830520 Free PMC article.
References
-
- Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75(2–3):291–295. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

