Accumulation of interspersed and sex-specific repeats in the non-recombining region of papaya sex chromosomes
- PMID: 24885930
- PMCID: PMC4035066
- DOI: 10.1186/1471-2164-15-335
Accumulation of interspersed and sex-specific repeats in the non-recombining region of papaya sex chromosomes
Abstract
Background: The papaya Y chromosome has undergone a degenerative expansion from its ancestral autosome, as a consequence of recombination suppression in the sex determining region of the sex chromosomes. The non-recombining feature led to the accumulation of repetitive sequences in the male- or hermaphrodite-specific regions of the Y or the Yh chromosome (MSY or HSY). Therefore, repeat composition and distribution in the sex determining region of papaya sex chromosomes would be informative to understand how these repetitive sequences might be involved in the early stages of sex chromosome evolution.
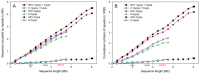
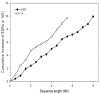
Results: Detailed composition of interspersed, sex-specific, and tandem repeats was analyzed from 8.1 megabases (Mb) HSY and 5.3 Mb corresponding X chromosomal regions. Approximately 77% of the HSY and 64% of the corresponding X region were occupied by repetitive sequences. Ty3-gypsy retrotransposons were the most abundant interspersed repeats in both regions. Comparative analysis of repetitive sequences between the sex determining region of papaya X chromosome and orthologous autosomal sequences of Vasconcellea monoica, a close relative of papaya lacking sex chromosomes, revealed distinctive differences in the accumulation of Ty3-Gypsy, suggesting that the evolution of the papaya sex determining region may accompany Ty3-Gypsy element accumulation. In total, 21 sex-specific repeats were identified from the sex determining region; 20 from the HSY and one from the X. Interestingly, most HSY-specific repeats were detected in two regions where the HSY expansion occurred, suggesting that the HSY expansion may result in the accumulation of sex-specific repeats or that HSY-specific repeats might play an important role in the HSY expansion. The analysis of simple sequence repeats (SSRs) revealed that longer SSRs were less abundant in the papaya sex determining region than the other chromosomal regions.
Conclusion: Major repetitive elements were Ty3-gypsy retrotransposons in both the HSY and the corresponding X. Accumulation of Ty3-Gypsy retrotransposons in the sex determining region of papaya X chromosome was significantly higher than that in the corresponding region of V. monoica, suggesting that Ty3-Gypsy could be crucial for the expansion and evolution of the sex determining region in papaya. Most sex-specific repeats were located in the two HSY expansion regions.
Figures



References
-
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. doi: 10.1007/BF02672069. - DOI
-
- Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, Salzberg SL, Feng L, Jones MR, Skelton RL, Murray JE, Chen C, Qian W, Shen J, Du P, Eustice M, Tong E, Tang H, Lyons E, Paull RE, Michael TP, Wall K, Rice DW, Albert H, Wang ML, Zhu YJ, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452(7190):991–996. doi: 10.1038/nature06856. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

