Real-time PCR assays for genotyping of Cryptococcus gattii in North America
- PMID: 24886039
- PMCID: PMC4032356
- DOI: 10.1186/1471-2180-14-125
Real-time PCR assays for genotyping of Cryptococcus gattii in North America
Abstract
Background: Cryptococcus gattii has been the cause of an ongoing outbreak starting in 1999 on Vancouver Island, British Columbia and spreading to mainland Canada and the US Pacific Northwest. In the course of the outbreak, C. gattii has been identified outside of its previously documented climate, habitat, and host disease. Genotyping of C. gattii is essential to understand the ecological and geographical expansion of this emerging pathogen.
Methods: We developed and validated a mismatch amplification mutation assay (MAMA) real-time PCR panel for genotyping C. gattii molecular types VGI-VGIV and VGII subtypes a,b,c. Subtype assays were designed based on whole-genome sequence of 20 C. gattii strains. Publically available multilocus sequence typing (MLST) data from a study of 202 strains was used for the molecular type (VGI-VGIV) assay design. All assays were validated across DNA from 112 strains of diverse international origin and sample types, including animal, environmental and human.
Results: Validation revealed each assay on the panel is 100% sensitive, specific and concordant with MLST. The assay panel can detect down to 0.5 picograms of template DNA.
Conclusions: The (MAMA) real-time PCR panel for C. gattii accurately typed a collection of 112 diverse strains and demonstrated high sensitivity. This is a time and cost efficient method of genotyping C. gattii best suited for application in large-scale epidemiological studies.
Figures
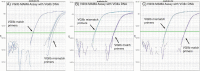
Similar articles
-
Molecular epidemiology reveals genetic diversity amongst isolates of the Cryptococcus neoformans/C. gattii species complex in Thailand.PLoS Negl Trop Dis. 2013 Jul 4;7(7):e2297. doi: 10.1371/journal.pntd.0002297. Print 2013. PLoS Negl Trop Dis. 2013. PMID: 23861989 Free PMC article.
-
Identification of novel hybrids between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII.Mycopathologia. 2012 Jun;173(5-6):337-46. doi: 10.1007/s11046-011-9491-x. Epub 2011 Nov 13. Mycopathologia. 2012. PMID: 22081254
-
Cryptococcus gattii in the Age of Whole-Genome Sequencing.mBio. 2015 Nov 17;6(6):e01761-15. doi: 10.1128/mBio.01761-15. mBio. 2015. PMID: 26578680 Free PMC article.
-
Cryptococcus gattii comparative genomics and transcriptomics: a NIH/NIAID White Paper.Mycopathologia. 2012 Jun;173(5-6):367-73. doi: 10.1007/s11046-011-9512-9. Epub 2011 Dec 17. Mycopathologia. 2012. PMID: 22179781 Review.
-
Cryptococcus gattii: a resurgent fungal pathogen.Trends Microbiol. 2011 Nov;19(11):564-71. doi: 10.1016/j.tim.2011.07.010. Epub 2011 Aug 29. Trends Microbiol. 2011. PMID: 21880492 Free PMC article. Review.
Cited by
-
Importance of Resolving Fungal Nomenclature: the Case of Multiple Pathogenic Species in the Cryptococcus Genus.mSphere. 2017 Aug 30;2(4):e00238-17. doi: 10.1128/mSphere.00238-17. eCollection 2017 Jul-Aug. mSphere. 2017. PMID: 28875175 Free PMC article.
-
Comparison of a Lateral Flow Assay and a Latex Agglutination Test for the Diagnosis of Cryptococcus Neoformans Infection.Curr Microbiol. 2021 Nov;78(11):3989-3995. doi: 10.1007/s00284-021-02664-w. Epub 2021 Sep 28. Curr Microbiol. 2021. PMID: 34581848 Free PMC article.
References
-
- D’Souza CA, Kronstad JW, Taylor G, Warren R, Yuen M, Hu G, Jung WH, Sham A, Kidd SE, Tangen K, Lee N, Zeilmaker T, Sawkins J, McVicker G, Shah S, Gnerre S, Griggs A, Zeng Q, Bartlett K, Li W, Wang X, Heitman J, Stajich JE, Fraser JA, Meyer W, Carter D, Schein J, Krzywinski M, Kwon-Chung KJ, Varma A. et al.Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. MBio. 2011;2:e00342–10. - PMC - PubMed
-
- Lockhart SR, Iqbal N, Bolden CB, DeBess EE, Marsden-Haug N, Worhle R, Thakur R, Harris JR. Epidemiologic cutoff values for triazole drugs in Cryptococcus gattii: correlation of molecular type and in vitro susceptibility. Diagn Microbiol Infect Dis. 2012;73(2):144–148. doi: 10.1016/j.diagmicrobio.2012.02.018. - DOI - PMC - PubMed
-
- Iqbal N, DeBess EE, Wohrle R, Sun B, Nett RJ, Ahlquist AM, Chiller T, Lockhart SR. Correlation of genotype and in vitro susceptibilities of Cryptococcus gattii strains from the Pacific Northwest of the United States. J Clin Microbiol. 2010;48(2):539–544. doi: 10.1128/JCM.01505-09. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

