Genomic analysis of the emergence of 20th century epidemic dysentery
- PMID: 24886041
- PMCID: PMC4038718
- DOI: 10.1186/1471-2164-15-355
Genomic analysis of the emergence of 20th century epidemic dysentery
Abstract
Background: Shigella dysenteriae type 1 (Sd1) causes recurrent epidemics of dysentery associated with high mortality in many regions of the world. Sd1 infects humans at very low infectious doses (10 CFU), and treatment is complicated by the rapid emergence of antibiotic resistant Sd1 strains. Sd1 is only detected in the context of human infections, and the circumstances under which epidemics emerge and regress remain unknown.
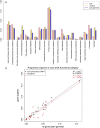
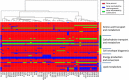
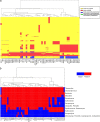
Results: Phylogenomic analyses of 56 isolates collected worldwide over the past 60 years indicate that the Sd1 clone responsible for the recent pandemics emerged at the turn of the 20th century, and that the two world wars likely played a pivotal role for its dissemination. Several lineages remain ubiquitous and their phylogeny indicates several recent intercontinental transfers. Our comparative genomics analysis reveals that isolates responsible for separate outbreaks, though closely related to one another, have independently accumulated antibiotic resistance genes, suggesting that there is little or no selection to retain these genes in-between outbreaks. The genomes appear to be subjected to genetic drift that affects a number of functions currently used by diagnostic tools to identify Sd1, which could lead to the potential failure of such tools.
Conclusions: Taken together, the Sd1 population structure and pattern of evolution suggest a recent emergence and a possible human carrier state that could play an important role in the epidemic pattern of infections of this human-specific pathogen. This analysis highlights the important role of whole-genome sequencing in studying pathogens for which epidemiological or laboratory investigations are particularly challenging.
Figures

Similar articles
-
Global phylogeography and evolutionary history of Shigella dysenteriae type 1.Nat Microbiol. 2016 Mar 21;1:16027. doi: 10.1038/nmicrobiol.2016.27. Nat Microbiol. 2016. PMID: 27572446
-
Genetic relatedness of ciprofloxacin-resistant Shigella dysenteriae type 1 strains isolated in south Asia.J Antimicrob Chemother. 2004 Oct;54(4):730-4. doi: 10.1093/jac/dkh425. Epub 2004 Sep 3. J Antimicrob Chemother. 2004. PMID: 15347639
-
Variation in the Shiga toxin region of 20th-century epidemic and endemic Shigella dysenteriae 1 strains.J Infect Dis. 2004 Jul 15;190(2):330-4. doi: 10.1086/421706. Epub 2004 Jun 9. J Infect Dis. 2004. PMID: 15216469
-
Using European travellers as an early alert to detect emerging pathogens in countries with limited laboratory resources.BMC Public Health. 2007 Jan 19;7:8. doi: 10.1186/1471-2458-7-8. BMC Public Health. 2007. PMID: 17239228 Free PMC article. Review.
-
Lethality of First Contact Dysentery Epidemics on Pacific Islands.Am J Trop Med Hyg. 2016 Aug 3;95(2):273-7. doi: 10.4269/ajtmh.16-0169. Epub 2016 May 16. Am J Trop Med Hyg. 2016. PMID: 27185765 Free PMC article. Review.
Cited by
-
Staphylococcus aureus Protein A Mediates Interspecies Interactions at the Cell Surface of Pseudomonas aeruginosa.mBio. 2016 May 24;7(3):e00538-16. doi: 10.1128/mBio.00538-16. mBio. 2016. PMID: 27222468 Free PMC article.
-
Global phylogeography and evolutionary history of Shigella dysenteriae type 1.Nat Microbiol. 2016 Mar 21;1:16027. doi: 10.1038/nmicrobiol.2016.27. Nat Microbiol. 2016. PMID: 27572446
-
The genomic signatures of Shigella evolution, adaptation and geographical spread.Nat Rev Microbiol. 2016 Apr;14(4):235-50. doi: 10.1038/nrmicro.2016.10. Epub 2016 Feb 29. Nat Rev Microbiol. 2016. PMID: 26923111 Review.
-
Looking Backward To Move Forward: the Utility of Sequencing Historical Bacterial Genomes.J Clin Microbiol. 2019 Jul 26;57(8):e00100-19. doi: 10.1128/JCM.00100-19. Print 2019 Aug. J Clin Microbiol. 2019. PMID: 31092597 Free PMC article. Review.
-
Genome sequencing, annotation and comparative genomic analysis of Shigella dysenteriae strain SD1D.Gut Pathog. 2014 Jul 11;6:28. doi: 10.1186/1757-4749-6-28. eCollection 2014. Gut Pathog. 2014. PMID: 25028600 Free PMC article.
References
-
- Niyogi SK. Shigellosis. J Microbiol. 2005;43(2):133–143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

