Viral metagenomic analysis of feces of wild small carnivores
- PMID: 24886057
- PMCID: PMC4030737
- DOI: 10.1186/1743-422X-11-89
Viral metagenomic analysis of feces of wild small carnivores
Abstract
Background: Recent studies have clearly demonstrated the enormous virus diversity that exists among wild animals. This exemplifies the required expansion of our knowledge of the virus diversity present in wildlife, as well as the potential transmission of these viruses to domestic animals or humans.
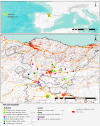
Methods: In the present study we evaluated the viral diversity of fecal samples (n = 42) collected from 10 different species of wild small carnivores inhabiting the northern part of Spain using random PCR in combination with next-generation sequencing. Samples were collected from American mink (Neovison vison), European mink (Mustela lutreola), European polecat (Mustela putorius), European pine marten (Martes martes), stone marten (Martes foina), Eurasian otter (Lutra lutra) and Eurasian badger (Meles meles) of the family of Mustelidae; common genet (Genetta genetta) of the family of Viverridae; red fox (Vulpes vulpes) of the family of Canidae and European wild cat (Felis silvestris) of the family of Felidae.
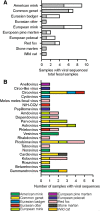
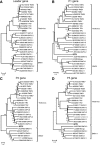
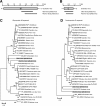
Results: A number of sequences of possible novel viruses or virus variants were detected, including a theilovirus, phleboviruses, an amdovirus, a kobuvirus and picobirnaviruses.
Conclusions: Using random PCR in combination with next generation sequencing, sequences of various novel viruses or virus variants were detected in fecal samples collected from Spanish carnivores. Detected novel viruses highlight the viral diversity that is present in fecal material of wild carnivores.
Figures


References
-
- Reusken CB, Haagmans BL, Müller MA, Gutierrez C, Godeke GJ, Meyer B, Muth D, Raj VS, Smits-De Vries L, Corman VM, Drexler JF, Smits SL, El Tahir YE, De Sousa R, van Beek J, Nowotny N, van Maanen K, Hidalgo-Hermoso E, Bosch BJ, Rottier P, Osterhaus A, Gortázar-Schmidt C, Drosten C, Koopmans MP. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. - DOI - PMC - PubMed
-
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q. et al.Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. - DOI - PubMed
-
- Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

