The large-scale investigation of gene expression in Leymus chinensis stigmas provides a valuable resource for understanding the mechanisms of poaceae self-incompatibility
- PMID: 24886329
- PMCID: PMC4045969
- DOI: 10.1186/1471-2164-15-399
The large-scale investigation of gene expression in Leymus chinensis stigmas provides a valuable resource for understanding the mechanisms of poaceae self-incompatibility
Abstract
Background: Many Poaceae species show a gametophytic self-incompatibility (GSI) system, which is controlled by at least two independent and multiallelic loci, S and Z. Until currently, the gene products for S and Z were unknown. Grass SI plant stigmas discriminate between pollen grains that land on its surface and support compatible pollen tube growth and penetration into the stigma, whereas recognizing incompatible pollen and thus inhibiting pollination behaviors. Leymus chinensis (Trin.) Tzvel. (sheepgrass) is a Poaceae SI species. A comprehensive analysis of sheepgrass stigma transcriptome may provide valuable information for understanding the mechanism of pollen-stigma interactions and grass SI.
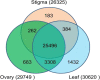
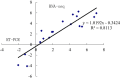
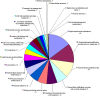
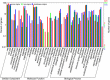
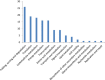
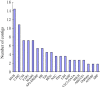
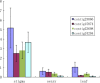
Results: The transcript abundance profiles of mature stigmas, mature ovaries and leaves were examined using high-throughput next generation sequencing technology. A comparative transcriptomic analysis of these tissues identified 1,025 specifically or preferentially expressed genes in sheepgrass stigmas. These genes contained a significant proportion of genes predicted to function in cell-cell communication and signal transduction. We identified 111 putative transcription factors (TFs) genes and the most abundant groups were MYB, C2H2, C3H, FAR1, MADS. Comparative analysis of the sheepgrass, rice and Arabidopsis stigma-specific or preferential datasets showed broad similarities and some differences in the proportion of genes in the Gene Ontology (GO) functional categories. Potential SI candidate genes identified in other grasses were also detected in the sheepgrass stigma-specific or preferential dataset. Quantitative real-time PCR experiments validated the expression pattern of stigma preferential genes including homologous grass SI candidate genes.
Conclusions: This study represents the first large-scale investigation of gene expression in the stigmas of an SI grass species. We uncovered many notable genes that are potentially involved in pollen-stigma interactions and SI mechanisms, including genes encoding receptor-like protein kinases (RLK), CBL (calcineurin B-like proteins) interacting protein kinases, calcium-dependent protein kinase, expansins, pectinesterase, peroxidases and various transcription factors. The availability of a pool of stigma-specific or preferential genes for L. chinensis offers an opportunity to elucidate the mechanisms of SI in Poaceae.
Figures

Similar articles
-
Transcriptomic Analysis Reveals a Comprehensive Calcium- and Phytohormone-Dominated Signaling Response in Leymus chinensis Self-Incompatibility.Int J Mol Sci. 2019 May 13;20(9):2356. doi: 10.3390/ijms20092356. Int J Mol Sci. 2019. PMID: 31085987 Free PMC article.
-
Identification of genes specifically or preferentially expressed in maize silk reveals similarity and diversity in transcript abundance of different dry stigmas.BMC Genomics. 2012 Jul 2;13:294. doi: 10.1186/1471-2164-13-294. BMC Genomics. 2012. PMID: 22748054 Free PMC article.
-
Identification and analysis of the stigma and embryo sac-preferential/specific genes in rice pistils.BMC Plant Biol. 2017 Mar 7;17(1):60. doi: 10.1186/s12870-017-1004-8. BMC Plant Biol. 2017. PMID: 28270108 Free PMC article.
-
How far are we from unravelling self-incompatibility in grasses?New Phytol. 2008;178(4):740-753. doi: 10.1111/j.1469-8137.2008.02421.x. Epub 2008 Mar 25. New Phytol. 2008. PMID: 18373516 Review.
-
Cell-cell signaling during the Brassicaceae self-incompatibility response.Trends Plant Sci. 2022 May;27(5):472-487. doi: 10.1016/j.tplants.2021.10.011. Epub 2021 Nov 27. Trends Plant Sci. 2022. PMID: 34848142 Review.
Cited by
-
TMT-Based Quantitative Proteomic Analysis Reveals the Crucial Biological Pathways Involved in Self-Incompatibility Responses in Camellia oleifera.Int J Mol Sci. 2020 Mar 14;21(6):1987. doi: 10.3390/ijms21061987. Int J Mol Sci. 2020. PMID: 32183315 Free PMC article.
-
Transcriptome and phytohormone analysis reveals a comprehensive phytohormone and pathogen defence response in pear self-/cross-pollination.Plant Cell Rep. 2017 Nov;36(11):1785-1799. doi: 10.1007/s00299-017-2194-0. Epub 2017 Sep 8. Plant Cell Rep. 2017. PMID: 28887590 Free PMC article.
-
Analysis on characteristics of female gametophyte and functional identification of genes related to inflorescences development of Kentucky bluegrass.Protoplasma. 2022 Jul;259(4):1061-1079. doi: 10.1007/s00709-021-01720-3. Epub 2021 Nov 6. Protoplasma. 2022. PMID: 34743240
-
Dissection of the style's response to pollination using transcriptome profiling in self-compatible (Solanum pimpinellifolium) and self-incompatible (Solanum chilense) tomato species.BMC Plant Biol. 2015 May 15;15:119. doi: 10.1186/s12870-015-0492-7. BMC Plant Biol. 2015. PMID: 25976872 Free PMC article.
-
New Insights on Drought Stress Response by Global Investigation of Gene Expression Changes in Sheepgrass (Leymus chinensis).Front Plant Sci. 2016 Jun 30;7:954. doi: 10.3389/fpls.2016.00954. eCollection 2016. Front Plant Sci. 2016. PMID: 27446180 Free PMC article.
References
-
- de Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. 2. Heidelberg: Springer; 2001.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

