Functional properties of the HIV-1 long terminal repeat containing single-nucleotide polymorphisms in Sp site III and CCAAT/enhancer binding protein site I
- PMID: 24886416
- PMCID: PMC4047001
- DOI: 10.1186/1743-422X-11-92
Functional properties of the HIV-1 long terminal repeat containing single-nucleotide polymorphisms in Sp site III and CCAAT/enhancer binding protein site I
Abstract
Background: HIV-1 gene expression is driven by the long terminal repeat (LTR), which contains many binding sites shown to interact with an array of host and viral factors. Selective pressures within the host as well as the low fidelity of reverse transcriptase lead to changes in the relative prevalence of genetic variants within the HIV-1 genome, including the LTR, resulting in viral quasispecies that can be differentially regulated and can potentially establish niches within specific cell types and tissues.
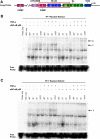
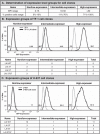
Methods: Utilizing flow cytometry and electromobility shift assays, specific single-nucleotide sequence polymorphisms (SNPs) were shown to alter both the phenotype of LTR-driven transcription and reactivation. Additional studies also demonstrated differential loading of transcription factors to probes derived from the double-variant LTR as compared to probes from the wild type.
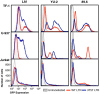
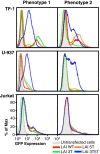
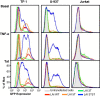
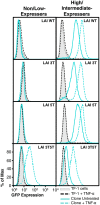
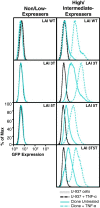
Results: This study has identified specific SNPs within CCAAT/enhancer binding protein (C/EBP) site I and Sp site III (3 T, C-to-T change at position 3, and 5 T, C-to-T change at position 5 of the binding site, respectively) that alter LTR-driven gene transcription and may alter the course of viral latency and reactivation. The HIV-1 LAI LTRs containing the SNPs of interest were coupled to a plasmid encoding green fluorescent protein (GFP), and polyclonal HIV-1 LTR-GFP stable cell lines utilizing bone marrow progenitor, T, and monocytic cell lines were constructed and utilized to explore the LTR phenotype associated with these genotypic changes.
Conclusions: Although the 3 T and 5 T SNPs have been shown to be low-affinity binding sites, the fact that they can still result in effective HIV-1 LTR-driven gene expression, particularly within the TF-1 cell line, has suggested that the low binding site affinities associated with the 3 T C/EBP site I and 5 T Sp site III are potentially compensated for by the interaction of nuclear factor-κB with its corresponding binding sites under selected physiological and cellular conditions. Additionally, tumor necrosis factor-α and Tat can enhance basal transcription of each SNP-specific HIV-1 LTR; however, differential regulation of the LTR is both SNP- and cell type-specific.
Figures
Similar articles
-
Region-specific distribution of human immunodeficiency virus type 1 long terminal repeats containing specific configurations of CCAAT/enhancer-binding protein site II in brains derived from demented and nondemented patients.J Neurovirol. 2004;10 Suppl 1:7-14. doi: 10.1080/753312746. J Neurovirol. 2004. PMID: 14982733
-
Impact of viral activators and epigenetic regulators on HIV-1 LTRs containing naturally occurring single nucleotide polymorphisms.Biomed Res Int. 2015;2015:320642. doi: 10.1155/2015/320642. Epub 2015 Jan 5. Biomed Res Int. 2015. PMID: 25629043 Free PMC article.
-
High-affinity interaction between HIV-1 Vpr and specific sequences that span the C/EBP and adjacent NF-kappaB sites within the HIV-1 LTR correlate with HIV-1-associated dementia.DNA Cell Biol. 2004 Apr;23(4):261-9. doi: 10.1089/104454904773819842. DNA Cell Biol. 2004. PMID: 15142383
-
DNA topoisomerase 1 represses HIV-1 promoter activity through its interaction with a guanine quadruplex present in the LTR sequence.Retrovirology. 2023 May 30;20(1):10. doi: 10.1186/s12977-023-00625-8. Retrovirology. 2023. PMID: 37254203 Free PMC article. Review.
-
HIV UTR, LTR, and Epigenetic Immunity.Viruses. 2022 May 18;14(5):1084. doi: 10.3390/v14051084. Viruses. 2022. PMID: 35632825 Free PMC article. Review.
Cited by
-
Evidence of Divergent Amino Acid Usage in Comparative Analyses of R5- and X4-Associated HIV-1 Vpr Sequences.Int J Genomics. 2017;2017:4081585. doi: 10.1155/2017/4081585. Epub 2017 May 17. Int J Genomics. 2017. PMID: 28620613 Free PMC article.
-
Targeting CCR5 as a Component of an HIV-1 Therapeutic Strategy.Front Immunol. 2022 Jan 20;12:816515. doi: 10.3389/fimmu.2021.816515. eCollection 2021. Front Immunol. 2022. PMID: 35126374 Free PMC article. Review.
-
Severe acute respiratory syndrome coronavirus 2 may exploit human transcription factors involved in retinoic acid and interferon-mediated response: a hypothesis supported by an in silico analysis.New Microbes New Infect. 2021 May;41:100853. doi: 10.1016/j.nmni.2021.100853. Epub 2021 Feb 27. New Microbes New Infect. 2021. PMID: 33680474 Free PMC article.
-
CNS-specific regulatory elements in brain-derived HIV-1 strains affect responses to latency-reversing agents with implications for cure strategies.Mol Psychiatry. 2016 Apr;21(4):574-84. doi: 10.1038/mp.2015.111. Epub 2015 Aug 25. Mol Psychiatry. 2016. PMID: 26303660 Free PMC article.
-
CRISPR-Cas9: A Preclinical and Clinical Perspective for the Treatment of Human Diseases.Mol Ther. 2021 Feb 3;29(2):571-586. doi: 10.1016/j.ymthe.2020.09.028. Epub 2020 Sep 20. Mol Ther. 2021. PMID: 33238136 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

