Hepatitis B virus X protein accelerates hepatocarcinogenesis with partner survivin through modulating miR-520b and HBXIP
- PMID: 24886421
- PMCID: PMC4046021
- DOI: 10.1186/1476-4598-13-128
Hepatitis B virus X protein accelerates hepatocarcinogenesis with partner survivin through modulating miR-520b and HBXIP
Abstract
Background: Hepatitis B virus X protein (HBx) plays crucial roles in hepatocarcinogenesis. However, the underlying mechanism remains elusive. We have reported that HBx is able to up-regulate survivin in hepatocellular carcinoma tissues. The oncopreotein hepatitis B X-interacting protein (HBXIP), a target of miR-520b, is involved in the development of cancer. In this study, we focus on the investigation of hepatocarcinogenesis mediated by HBx.
Methods: The expression of HBx and survivin was examined in the liver tissues of HBx-Tg mice. The effect of HBx/survivin on the growth of LO2-X-S cells was determined by colony formation and transplantation in nude mice. The effect of HBx/survivin on promoter of miR-520b was determined by Western blot analysis, luciferase reporter gene assay, co-immunoprecipitation (co-IP) and chromatin immunoprecipitation (ChIP), respectively. The expression of HBx, survivin and HBXIP was detected by immunohistochemistry and real-time PCR in clinical HCC tissues, respectively. The DNA demethylation of HBXIP promoter was examined. The functional influence of miR-520b and HBXIP on proliferation of hepatoma cells was analyzed by MTT, colony formation, EdU and transplantation in nude mice in vitro and in vivo.
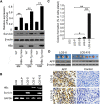
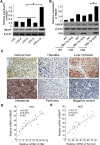
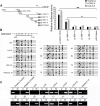
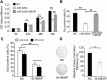
Results: In this study, we provided evidence that HBx up-regulated survivin in the liver cancer tissues of HBx-Tg mice aged 18 M. The engineered LO2 cell lines with survivin and/or HBx were successfully established, termed LO2-X-S. MiR-520b was down-regulated in LO2-X-S cells and clinical HCC tissues. Our data revealed that HBx survivin-dependently down-regulated miR-520b through interacting with Sp1 in the cells. HBXIP was highly expressed in LO2-X-S cells, liver cancer tissues of HBx-Tg mice aged 18 M and clinical HCC tissues (75.17%, 112/149). The expression level of HBXIP was positively associated with those of HBx or survivin in clinical HCC tissues. In addition, we showed that HBx survivin-dependently up-regulated HBXIP through inducing demethylation of HBXIP promoter in LO2-X-S cells and clinical HCC tissues. In function, low level miR-520b and high level HBXIP mediated by HBx with partner survivin contributed to the growth of LO2-X-S cells in vitro and in vivo.
Conclusion: HBx accelerates hepatocarcinogenesis with partner survivin through modulating tumor suppressor miR-520b and oncoprotein HBXIP.
Figures

References
-
- Peng Z, Zhang Y, Gu W, Wang Z, Li D, Zhang F, Qiu G, Xie K. Integration of the hepatitis B virus X fragment in hepatocellular carcinoma and its effects on the expression of multiple molecules: a key to the cell cycle and apoptosis. Int J Oncol. 2005;26:467–473. - PubMed
-
- Yun C, Cho H, Kim SJ, Lee JH, Park SY, Chan GK. Mitotic aberration coupled with centrosome amplification is induced by hepatitis B virus X oncoprotein via the Ras-mitogen-activated protein/extracellular signal-regulated kinase-mitogen-activated protein pathway. Mol Cancer Res. 2004;2:159–169. - PubMed
-
- You X, Liu F, Zhang T, Lv N, Liu Q, Shan C, Du Y, Kong G, Wang T, Ye L, Zhang X. Hepatitis B virus X protein upregulates Lin28A/Lin28B through Sp-1/c-Myc to enhance the proliferation of hepatoma cells. Oncogene. 2013;33:449–460. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

