MP-Align: alignment of metabolic pathways
- PMID: 24886436
- PMCID: PMC4045882
- DOI: 10.1186/1752-0509-8-58
MP-Align: alignment of metabolic pathways
Abstract
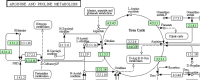
Background: Comparing the metabolic pathways of different species is useful for understanding metabolic functions and can help in studying diseases and engineering drugs. Several comparison techniques for metabolic pathways have been introduced in the literature as a first attempt in this direction. The approaches are based on some simplified representation of metabolic pathways and on a related definition of a similarity score (or distance measure) between two pathways. More recent comparative research focuses on alignment techniques that can identify similar parts between pathways.
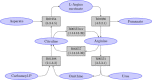
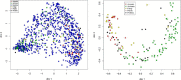
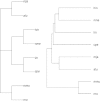
Results: We propose a methodology for the pairwise comparison and alignment of metabolic pathways that aims at providing the largest conserved substructure of the pathways under consideration. The proposed methodology has been implemented in a tool called MP-Align, which has been used to perform several validation tests. The results showed that our similarity score makes it possible to discriminate between different domains and to reconstruct a meaningful phylogeny from metabolic data. The results further demonstrate that our alignment algorithm correctly identifies subpathways sharing a common biological function.
Conclusion: The results of the validation tests performed with MP-Align are encouraging. A comparison with another proposal in the literature showed that our alignment algorithm is particularly well-suited to finding the largest conserved subpathway of the pathways under examination.
Figures


Similar articles
-
Functional Alignment of Metabolic Networks.J Comput Biol. 2016 May;23(5):390-9. doi: 10.1089/cmb.2015.0203. Epub 2016 Jan 13. J Comput Biol. 2016. PMID: 26759932
-
The Application of the Weighted k-Partite Graph Problem to the Multiple Alignment for Metabolic Pathways.J Comput Biol. 2017 Dec;24(12):1195-1211. doi: 10.1089/cmb.2017.0064. Epub 2017 Sep 11. J Comput Biol. 2017. PMID: 28891687
-
C-GRAAL: common-neighbors-based global GRAph ALignment of biological networks.Integr Biol (Camb). 2012 Jul;4(7):734-43. doi: 10.1039/c2ib00140c. Epub 2012 Jan 10. Integr Biol (Camb). 2012. PMID: 22234340
-
Grohar: Automated Visualization of Genome-Scale Metabolic Models and Their Pathways.J Comput Biol. 2018 May;25(5):505-508. doi: 10.1089/cmb.2017.0209. Epub 2018 Feb 20. J Comput Biol. 2018. PMID: 29461874
-
Comparing methods for metabolic network analysis and an application to metabolic engineering.Gene. 2013 May 25;521(1):1-14. doi: 10.1016/j.gene.2013.03.017. Epub 2013 Mar 26. Gene. 2013. PMID: 23537990 Review.
Cited by
-
Reconstructing Phylogeny by Aligning Multiple Metabolic Pathways Using Functional Module Mapping.Molecules. 2018 Feb 23;23(2):486. doi: 10.3390/molecules23020486. Molecules. 2018. PMID: 29473850 Free PMC article.
-
Low-Cost Algorithms for Metabolic Pathway Pairwise Comparison.Biomimetics (Basel). 2022 Feb 21;7(1):27. doi: 10.3390/biomimetics7010027. Biomimetics (Basel). 2022. PMID: 35225919 Free PMC article.
-
Metadag: a web tool to generate and analyse metabolic networks.BMC Bioinformatics. 2025 Jan 28;26(1):31. doi: 10.1186/s12859-025-06048-w. BMC Bioinformatics. 2025. PMID: 39875845 Free PMC article.
-
The alignment of enzymatic steps reveals similar metabolic pathways and probable recruitment events in Gammaproteobacteria.BMC Genomics. 2015 Nov 17;16:957. doi: 10.1186/s12864-015-2113-0. BMC Genomics. 2015. PMID: 26578309 Free PMC article.
-
MetNet: A two-level approach to reconstructing and comparing metabolic networks.PLoS One. 2021 Feb 12;16(2):e0246962. doi: 10.1371/journal.pone.0246962. eCollection 2021. PLoS One. 2021. PMID: 33577575 Free PMC article.
References
-
- KEGG pathway database - Kyoto University Bioinformatics Centre. [ http://www.genome.jp/kegg/pathway.html]
-
- BioModels Database. [ http://www.ebi.ac.uk/biomodels]
-
- MetaCyc Encyclopedia of Metabolic Pathways. [ http://metacyc.org]
-
- Sridhar P, Kahveci T, Ranka S. An iterative algorithm for metabolic network-based drug target identification. Pac Symp Biocomput. 2007;12:88–99. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

