Alcohol induces cell proliferation via hypermethylation of ADHFE1 in colorectal cancer cells
- PMID: 24886599
- PMCID: PMC4057807
- DOI: 10.1186/1471-2407-14-377
Alcohol induces cell proliferation via hypermethylation of ADHFE1 in colorectal cancer cells
Abstract
Background: The hypermethylation of Alcohol dehydrogenase iron containing 1 (ADHFE1) was recently reported to be associated with colorectal cancer (CRC) differentiation. However, the effect of alcohol on ADHFE1 hypermethylation in CRC is still unclear.
Methods: The methylation status and expression levels of ADHFE1 were investigated in primary tumor tissues and adjacent normal tissues of 73 patients with CRC, one normal colon cell line, and 4 CRC cell lines (HT-29, SW480, DLD-1, and LoVo) by quantitative methylation-specific polymerase chain reaction (QMSP) and real-time reverse transcription polymerase chain reaction (real time PCR), respectively. The effect of alcohol on the methylation status of ADHFE1 was analyzed in HT-29, SW480, DLD-1, and CCD18Co cells using QMSP, real-time PCR, immunoblot, and cell proliferation assay.
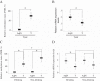
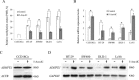
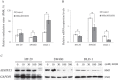
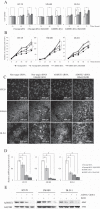
Results: ADHFE1 was hypermethylated in 69 of 73 CRC tissues (95%) compared to adjacent normal tissues (p<0.05). The mRNA expression of ADHFE1 was significantly reduced in CRC compared to adjacent normal tissues (p<0.05) and its expression was decreased in the alcohol consumption group (p<0.05). ADHFE1 was hypermethylated and its expression was decreased in 4 CRC cell lines compared with normal colon cell line. Alcohol induced hypermethylation of ADHFE1, decreased its expression, and stimulated cell proliferation of HT-29, SW480, and DLD-1cells.
Conclusion: These results demonstrate that the promoter hypermethylation of ADHFE1 is frequently present in CRC and alcohol induces methylation-mediated down expression of ADHFE1 and proliferation of CRC cells.
Figures
Similar articles
-
Alcohol dehydrogenase, iron containing, 1 promoter hypermethylation associated with colorectal cancer differentiation.BMC Cancer. 2013 Mar 22;13:142. doi: 10.1186/1471-2407-13-142. BMC Cancer. 2013. PMID: 23517143 Free PMC article.
-
Promoter hypermethylation of membrane type 3 matrix metalloproteinase is associated with cell migration in colorectal adenocarcinoma.Cancer Genet. 2015 May;208(5):261-70. doi: 10.1016/j.cancergen.2015.04.009. Epub 2015 May 1. Cancer Genet. 2015. PMID: 26002729
-
Demethylation of RUNX3 by vincristine in colorectal adenocarcinoma cells.Anticancer Res. 2014 Jan;34(1):133-40. Anticancer Res. 2014. PMID: 24403453
-
Hypermethylation Of ADHFE1 Promotes The Proliferation Of Colorectal Cancer Cell Via Modulating Cell Cycle Progression.Onco Targets Ther. 2019 Oct 4;12:8105-8115. doi: 10.2147/OTT.S223423. eCollection 2019. Onco Targets Ther. 2019. PMID: 31632063 Free PMC article.
-
NGX6 gene mediated by promoter methylation as a potential molecular marker in colorectal cancer.BMC Cancer. 2010 Apr 27;10:160. doi: 10.1186/1471-2407-10-160. BMC Cancer. 2010. PMID: 20423473 Free PMC article.
Cited by
-
Genome-wide DNA methylation profiles of low- and high-grade adenoma reveals potential biomarkers for early detection of colorectal carcinoma.Clin Epigenetics. 2020 Apr 21;12(1):56. doi: 10.1186/s13148-020-00851-3. Clin Epigenetics. 2020. PMID: 32317010 Free PMC article.
-
The incidence and risk factors of sessile serrated adenomas in left side colon cancer patients after curative surgery.Medicine (Baltimore). 2020 Jul 17;99(29):e20799. doi: 10.1097/MD.0000000000020799. Medicine (Baltimore). 2020. PMID: 32702823 Free PMC article.
-
Epigenome-Wide DNA Methylation Profiling in Colorectal Cancer and Normal Adjacent Colon Using Infinium Human Methylation 450K.Diagnostics (Basel). 2022 Jan 14;12(1):198. doi: 10.3390/diagnostics12010198. Diagnostics (Basel). 2022. PMID: 35054365 Free PMC article.
-
Identifying CpG sites with different differential methylation frequencies in colorectal cancer tissues based on individualized differential methylation analysis.Oncotarget. 2017 Jul 18;8(29):47356-47364. doi: 10.18632/oncotarget.17647. Oncotarget. 2017. PMID: 28537885 Free PMC article.
-
Using Comorbidity Pattern Analysis to Detect Reliable Methylated Genes in Colorectal Cancer Verified by Stool DNA Test.Genes (Basel). 2021 Sep 28;12(10):1539. doi: 10.3390/genes12101539. Genes (Basel). 2021. Retraction in: Genes (Basel). 2024 Jun 19;15(6):805. doi: 10.3390/genes15060805. PMID: 34680934 Free PMC article. Retracted.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

